Figures & data
Fig. 1 (Colour online) (A) Oil palm seen with typical symptoms of orange spotting with severe crown discolouration. (B) A closer view of the affected fronds with orange spotting symptoms. (Figures courtesy of Malaysian Palm Oil Board.)

Table 1. Comparison of the different RNA extraction methods used in this study.
Fig. 2 RT-PCR product from symptomatic samples extracted using the CTAB extraction and NETME methods. Lane M: 100 bp ladder; Lanes 1–3: symptomatic samples extracted using CTAB; Lane 4: SB 346 extracted using CTAB; Lane 5–7: symptomatic tissues extracted using NETME; Lane 8: SB 346 extracted using NETME; Lane 9: negative control (no template).
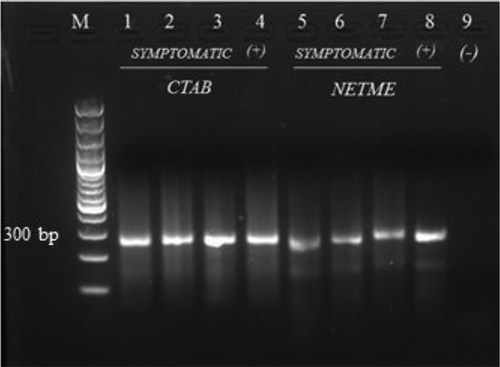
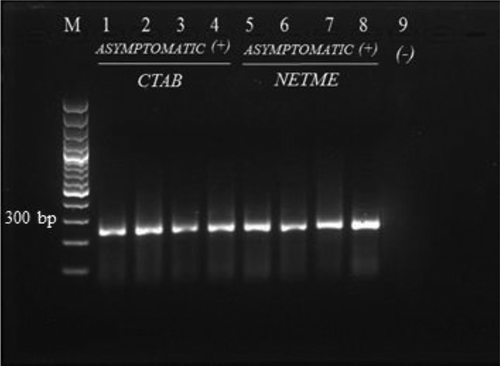
Fig. 3 RT-PCR product from asymptomatic samples extracted using the CTAB extraction and NETME methods. Lane M: 100 bp ladder; Lanes 1–3: asymptomatic samples extracted using CTAB; Lane 4: SB 346 extracted using CTAB; Lane 5–7: asymptomatic sample extracted using NETME; Lane 8: SB 346 extracted using NETME; Lane 9: negative control (no template).