Figures & data
Table 1. Oligonucleotide primers used in this study.
Fig. 1 Adhesion of sand particles on hyphae and mycelia of Fusarium graminearum in a liquid medium supplemented with sand. a, b, The medium was incubated at 150 rpm in 30°C for 48 h after inoculated with F. graminearum conidia (a) and mycelia (b). c, The mycelia in (b) after washing with 40 mL of water. Bars = 10 μm.
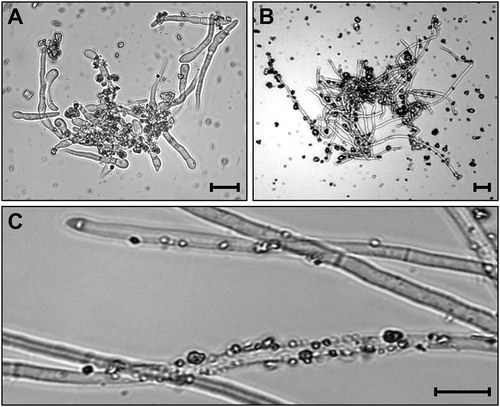
Table 2. Quantity and range of the 260/280 ratio for genomic DNA extracted from mycelia.
Table 3. Calculated possibilities (p-values) of a two-way ANOVA to investigate the variations in DNA quantity.
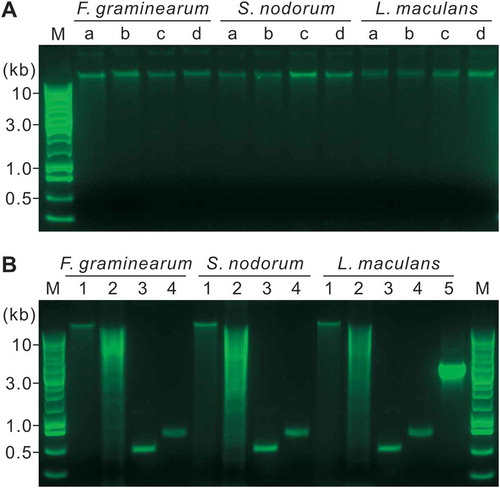
Fig. 2 (Colour online) Quality assessment of the DNA extracted by the APHL protocol on 1% agarose gels. M, Promega 1-kb DNA ladder. a, Five-hundred ng of DNA in a volume of 10 μL was analysed. The DNA was extracted from mycelia derived from YPG media inoculated with conidia (a) or mycelia (b), or YPG/sand inoculated with conidia (c) or mycelia (d). b, The DNA in (c) were further analysed. 1, Three-hundred ng of DNA in a volume of 10 μL were incubated at 37°C for 2.5 h; 2, Three μg DNA in a volume of 50 μL were digested with Hind III at 37°C for 2.5 h; 3–5, PCR products from 50 ng DNA templates using the primer pair ITS1/ITS4 (3), H729/H730 (4) or 13059/13062 (5).