Figures & data
Fig. 1 (Colour online) Symptoms of necrosis and yellowing in older leaves observed in palms surveyed in Grand-Lahou corresponding to species B. aethiopium (A, old plant; B, young plant); E. guineensis (C); and R. vinifera (E); and CILY-like stage 2 and 3 symptoms in C. nucifera (D).

Table 1. Results of PCR testing of palm species E. guineensis, B. aethiopium, R. vinifera and C. nucifera surveyed in Grand-Lahou for the presence of phytoplasma DNA.
Table 2. Sequence similarities based on the 16S rRNA gene sequences of the 16SrXXII-C phytoplasma and phytoplasma strains in 16SrXXII and 16SrIV groups/subgroups.
Fig. 2 Virtual Sau3AI profile obtained from the iPhyClassifier analysis of the R16F2n/R2 sequences from palms and 16SrXXII representatives. Lanes MW: Molecular weight marker (NEB 100 bp DNA Ladder), Lane 1: R. vinifera (GenBank accession number, AC: KY711302), representing the same profile for B. aethiopium (AC: KY711392), E. guineensis (AC: KY711391) and C. nucifera (AC: KY767914), Lane 2: LYM phytoplasma (AC: KP938848, 16SrXXII-A); Lane 3: CILY phytoplasma (AC: KC999037, 16SrXXII-B).
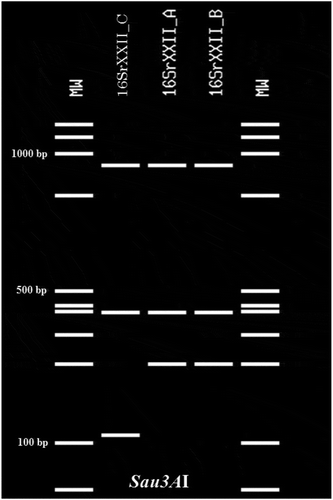
Fig. 3 Sau3AI profile obtained from the R16F2n/R2 amplicons of the phytoplasmas identified in E. guineensis, B. aethiopium, R. vinifera and C. nucifera palms, and positive controls of subgroups 16SrXXII-A (LYM), and 16SrXXII-B (CSPWD, CILY). Lane M: Molecular weight marker (Quick Load 100 bp DNA Ladder, New England BioLabs); Lane 1, R. vinifera (16SrXXII-C); Lane 2, B. aethiopium (16SrXXII-C); Lane 3, E. guineensis (16SrXXII-C); Lane 4, C. nucifera (16SrXXII-C); Lane 5, CILY phytoplasma (16SrXXII-B, Côte d’Ivoire); Lane 6, CSPWD phytoplasma (16SrXXII-B, Ghana); Lane 7, LYM phytoplasma (16SrXXII-A).
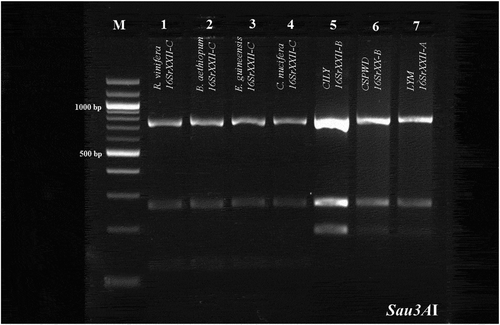
Fig. 4 Virtual (a) and actual (b) HpyCH4III profiles obtained from the SecA amplicons of the phytoplasmas identified in E. guineensis, B. aethiopium, R. vinifera and C. nucifera palms, and phytoplasma strains in subgroups 16SrXXII-A (LYM), and 16SrXXII-B (CSPWD, CILY). Lanes MW (A): NEB 100 bp DNA ladder. Lane M (B): Molecular weight marker (Quick Load 100 bp DNA Ladder, New England BioLabs); Lanes 1, R. vinifera (16SrXXII-C); Lanes 2, B. aethiopium (16SrXXII-C); Lanes 3, E. guineensis (16SrXXII-C); Lanes 4, C. nucifera (16SrXXII-C); Lanes 5, CILY phytoplasma (16SrXXII-B, Côte d’Ivoire); Lanes 6, CSPWD phytoplasma (16SrXXII-B, Ghana); Lanes 7, LYM phytoplasma (16SrXXII-A).
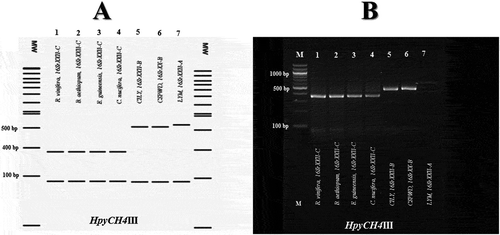
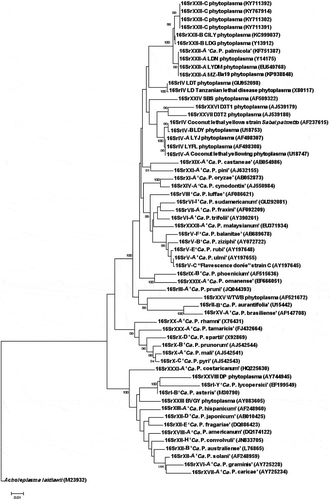
Fig. 5 Phylogenetic tree based on the R16F2n/R2 sequences of the 16SrXXII-C phytoplasmas and selected 16Sr phytoplasma groups. ‘Ca. P. sp.’: ‘Candidatus Phytoplasma sp.’; CILY: Côte d’Ivoire Lethal Yellowing; CSPWD, Cape St. Paul Wilt Disease; LDG: Lethal Decline (Ghana); LYDM: Lethal Yellowing Disease (Mozambique); LDN: Lethal Decline (Nigeria); LDT: Lethal Decline (Tanzania); SBS: Sorghum Bunchy Shoot; D3T1 and D3T2: phytoplasmas from sugarcane; C. palmata: Carludovica palmata; LDY: Lethal Decline (Yucatan); LYFL: Lethal Yellowing (Florida); LYJ: Lethal Yellowing (Jamaica); BVGY: Buckland Valley Grapevine Yellows; A. laidlawii, outgroup. GenBank accession numbers shown above branches. Bootstrap values shown are greater than 85%. Branch lengths are proportional to the number of inferred state transformations. Bar indicates 0.01 substitutions per nucleotide position.