Figures & data
Table 1. List of Canadian ‘Pacific Gala’ apple samples examined in this study with positive results.
Fig. 1 (Colour online) Swelling and radial limb cracking on a symptomatic ‘Pacific Gala’ apple tree.

Fig. 2 (Colour online) Agarose gel analysis of RT-PCR products stained with EtBr. The RT-PCR products were generated using primer pair AHVd-13F_PG & AHVd-12R_PG (a) and AHVd-88F_PG & AHVd-87R_PG (b). m = 100 bp Invitrogen ladder, 1 = PG_NS15_05 (symptomatic), 2 = PG_OK15_05 (symptomatic), 3 = PG_OK15_07 (asymptomatic), 4 = AP_SD15_ASS1, 5 = RT-PCR negative control.
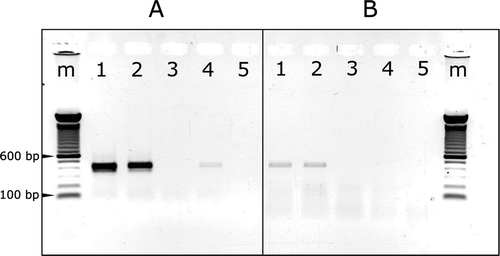
Table 2. Pairwise comparisons of nucleotide sequences within and between the variants of apple hammerhead viroid-like RNA detected in ‘Fuji’ apple (KR605 506 is the reference variant from the Chinese isolate) and the isolates of ‘Pacific Gala’ apple from Nova Scotia (NS) and the Okanagan (OK) Canada. Numbers below the diagonal show the number of nucleotides that differed for each comparison and identities (%) are shown above the diagonal. The data were obtained using MEGA6 (Tamura et al. Citation2013).
Table 3. AU/GC content of representative sequences for AHVd RNA-PG (NS15-5) and AHVd RNA-F (KR605506). Subsections of the entire sequence relating to predicted secondary structures are also treated individually. Analyses were carried out using Clone Manager 9 (Sci-Ed, Denver).
Table 4. Locations of polymorphic nucleotides (underlined) in AHVd RNA-PG and AHVd RNA-F variants.
Fig. 3 Bayesian phylogenetic tree illustrating the relationships between isolates of AHVd RNA-PG (OK = Okanagan, NS = Nova Scotia) and AHVd RNA-F variants. AHVd RNA-F variants are indicated as KR605506 and AHVd in the tree. Posterior probabilities of 70 per cent or higher are shown at group nodes. Scale bars indicate the number of substitutions per residue.
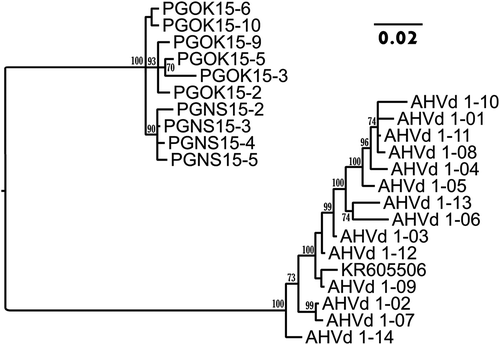
Fig. 4 (Colour online) Nucleotide sequence and predicted minimum free-energy secondary structures for AHVd RNA-F representative variant KR605506 (a) and AHVd RNA-PG representative isolate NS15-5 (b), predicted using RNAfold (Hofacker Citation2003). Filled and open symbols pertain respectively to the plus and minus strands. Residues conserved in hammerhead structures of viroid and viroid-like satellite RNAs are shown with bars, and the inferred self-cleavage sites are identified with arrowheads. The hammerhead region itself is flanked by hammer icons, and thin arrows denote the locations of abutting primer pairs used in amplification. (c) Schematic representation of the proposed hammerhead structures of AHVd RNA-PG. Helices I-III and loops L1/L2 are indicated, and several strictly conserved nucleotides are boxed. Icons indicate the self-cleavage sites.
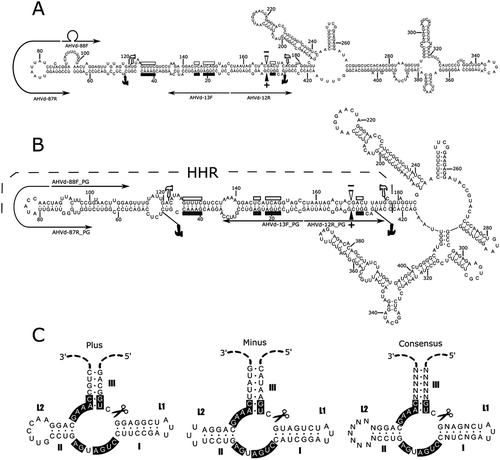
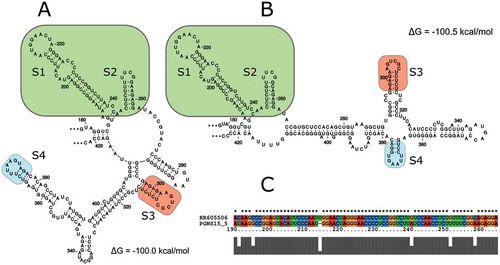
Fig. 5 (Colour online) Comparison of the nucleotide sequence and predicted secondary structures of the ‘branched’ moieties in the loop rich regions (LRRs) of AHVd RNA sequences of the AHVd RNA-PG representative isolate NS15-5 (a) and the AHVd RNA-F representative variant KR605506 (b). Nucleotides 179–422 and 178–424 were used in an independent RNAfold analysis with analogous stem/loop structures indicated (S1–S4). An alignment comparing the portion of (a) and (b) found in the green box (S1 and S2) is shown in (c). Minimum free energies are also shown.