Figures & data
Table 1. Primers used for PCR amplification and sequencing in this study.
Fig. 1 (Colour online) Symptoms of anthracnose on passion fruit and morphological features of the pathogenic fungus Colletotrichum brevisporum (teleomorph: Glomerella sp.). (a–d) Symptoms of anthracnose on stems (a), tendrils (b), fruits (c), and leaves (d) in the field. (e–f) Purified upper colony (e) and reverse colony (f) of C. brevisporum grown on PDA. (g–h) conidia (g) and conidial appressoria (h) (scale bar = 20 μm). (i–j) acervuli (i) and perithecia (j) (scale bar = 50 μm). (k) asci (scale bar = 40 μm). (l) ascospores (scale bar = 20 μm). (m–p) symptoms observed on fruits (m), stems (n), tendrils (o) and leaves (p) 14 days after inoculation with a conidial suspension of C. brevisporum.

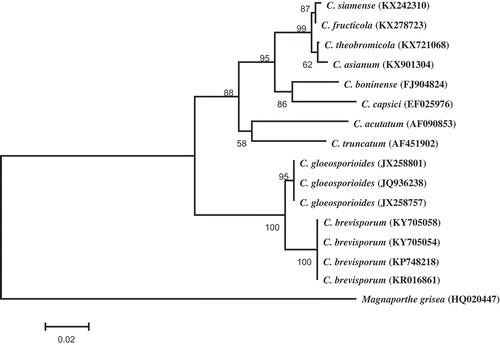
Fig. 2 Phylogenetic tree obtained through Neighbour-joining method using MEGA 5.1 based on ITS rDNA sequences of two representative isolates C. brevisporum (KY705054 and KY705058), and 13 Colletotrichum spp. isolates retrieved from GenBank. The numbers above the branches indicate bootstrap values resulting from 1000 replicates. Magnaporthe grisea was used as an outgroup.