Figures & data
Fig. 1 (Colour online) a, b, c, Field symptoms of leaf spot disease on cattleya orchid (Cattleya lueddemanniana var. lueddemanniana) caused by Neoscytalidium orchidacearum. d, Leaf spot disease of orchid inoculated with N. orchidacearum CMU287 (left) and control (right). e, Colonies of N. orchidacearum CMU287 from the top (left) and bottom (right) on PDA at 25°C after 1 week. f, Arthric chains of conidia growing on PDA. g, Conidiomata formed on pine needles in culture. h, i, j, k, Conidia of coelomycetous state. Scale bars: A = 50 mm; b, c, d and e = 10 mm; f = 10 µm; g = 50 µm; h, i, j and k = 3 µm.
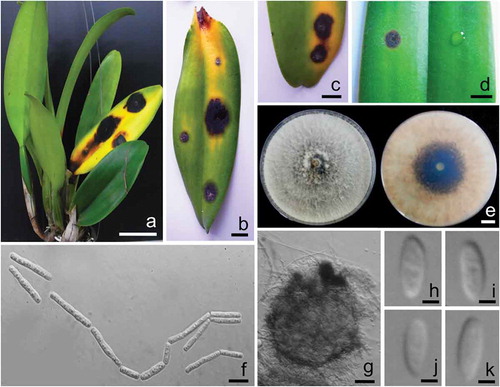
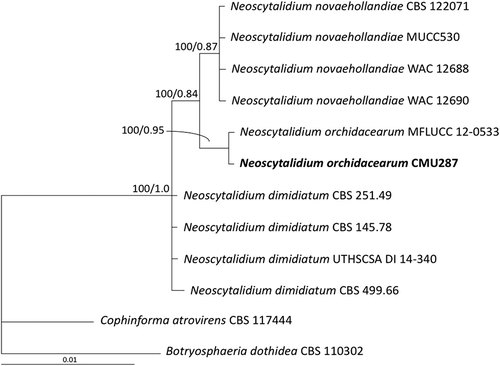
Fig. 2 Phylogram derived from maximum likelihood analysis of combined internal transcribed spacer and large subunit of rDNA sequence data for 12 fungal strains including isolate CMU287 isolated from diseased orchid leaf tissue in this study. The sequences for the other 11 fungal strains were obtained from the GenBank database. Botryosphaeria dothidea and Cophinforma atrovirens were used as outgroups. The numbers above branches represent maximum likelihood bootstrap percentages (left) and Bayesian posterior probabilities (right). Only bootstrap values ≥ 50% are shown. Isolate CMU287 sequenced in this study is indicated in bold.