Figures & data
Fig. 1 (Colour online) Distribution of bacterial leaf spot caused by Xanthomonas arboricola pv. pruni in China. The shaded areas are where peach samples were collected in this study.
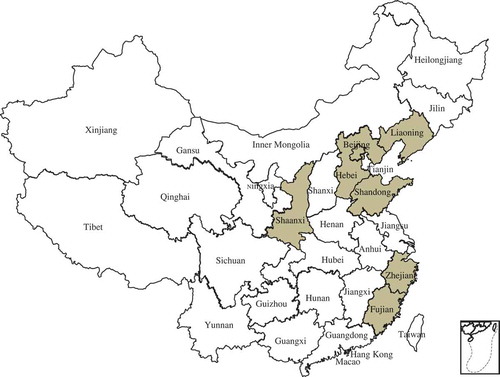
Fig. 2 (Colour online) Typical symptoms of bacterial leaf spot disease on naturally infected peach leaves, twigs and fruits. (a) brown and perforated symptomatic peach leaf collected from Beijing Pinggu; (b) severe symptoms on the entire peach leaf collected from Hebei Changli; (c) dark canker symptomatic peach twig collected from Beijing Pinggu and (d, e) severe brown, circular, water soaked spots and cracking symptoms observed on peach fruit collected from Beijing Pinggu district.
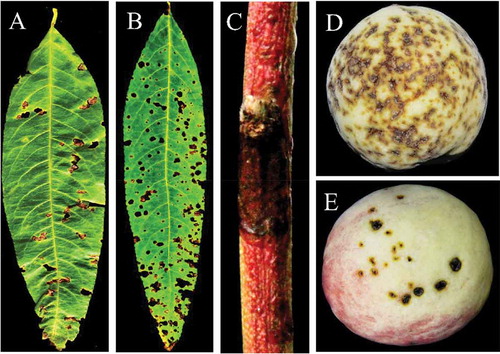
Fig. 3 (Colour online) Leaf spot disease isolates from symptomatic peach samples on Nutrient Agar (a) and Sucrose Peptone Agar (b) after 3 days of incubation at 28°C.
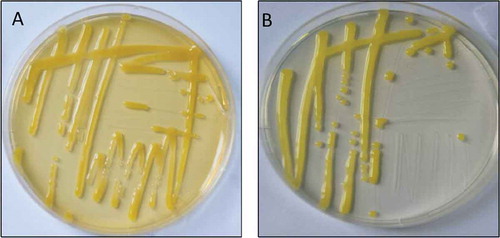
Fig. 4 (a) PCR amplification fragments of 16S rRNA region from isolates of Xanthomonas arboricola pv. pruni. M: DL2000 DNA marker (TIANGEN, Beijing, China). Lane NC: Negative control. Lanes 1–11: isolates of Xanthomonas arboricola pv. pruni. (b) Detection results of Xanthomonas arboricola pv. pruni by PCR using primer pairs of XapY17-F and XapY-R from naturally infected and greenhouse inoculated peach samples and 3-day old pure culture media. Lane M: DL2000 DNA marker (Tiangen, Beijing, China). Lanes 1–6: isolates of Xanthomonas arboricola pv. pruni from field grown peach samples. W: water control: H: DNA from healthy peach samples. Lanes 7–12: isolates of Xanthomonas arboricola pv. pruni from greenhouse inoculated peach seedlings.
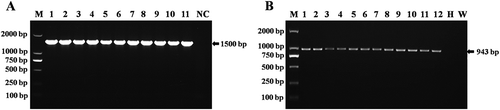
Fig. 5 (Colour online) (a) Hypersensitive response (HR) by Xap isolates on tobacco leaf (Nicotiana benthamiana). (b) Xap isolates inoculated and their symptoms 10 days after inoculation on peach seedlings.
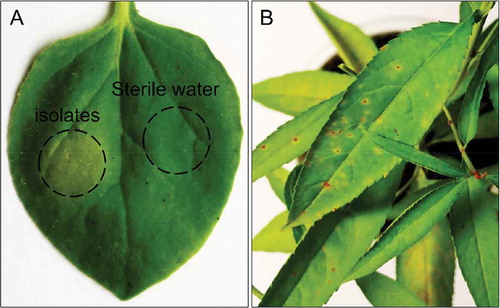
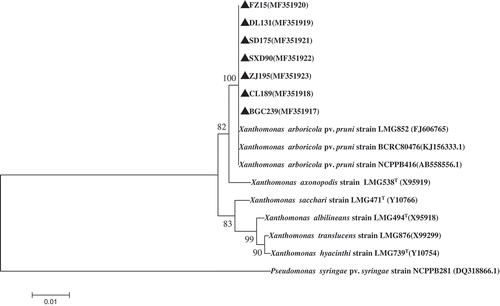
Fig. 6 Phylogenetic tree constructed with the 16S rRNA region sequence of Xanthomonas arboricola pv. pruni isolated in this study (MF351917–MF351923) and other species of Xanthomonads retrieved from GenBank. The tree was constructed by neighbour-joining method from the alignments of 16S rRNA sequences from this study and representative Xanthomonas species from NCBI using MEGA software (version 6) using 1000 bootstrap replicates; only bootstrap values greater than 50% are indicated . Pseudomonas syringae pv. syringae (accession no. DQ318866.1) was used as the out-group taxon.