Figures & data
Fig. 1 (Colour online) Colonies, conidia, and sporulation patterns of three Alternaria species after 7 days of growth on PDA or PCA plates. a–c, Colonies of isolates representing A. tenuissima, A. alternata, A. solani on PDA plates. d–f, Conidia of isolates representing A. tenuissima, A. alternata, A. solani on PCA plates, scale bars: 50 μm. g–h, sporulation patterns of isolates representing A. tenuissima, A. alternata on PCA plates, scale bars: 200 μm.
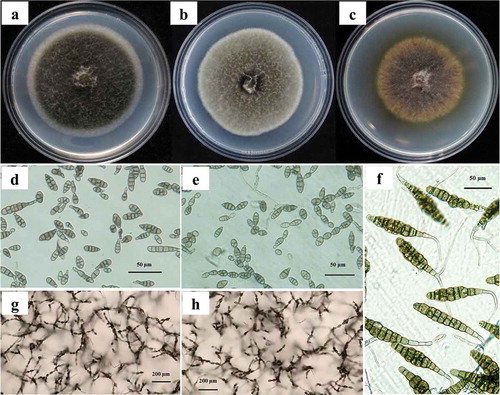
Fig. 2 (Colour online) Pathogenicity of the representative isolates of three Alternaria species on detached potato leaflets. a-b, sterile water controls. c–d, A. tenuissima. e–f, A. alternata. g–h, A. solani. The experiment was conducted using detached apical leaflets from 45-day-old plants of potato cv. Favorite. A spore suspension of 106 conidia·ml−1 was inoculated on the upper surface of each leaflet (one point per leaflet).
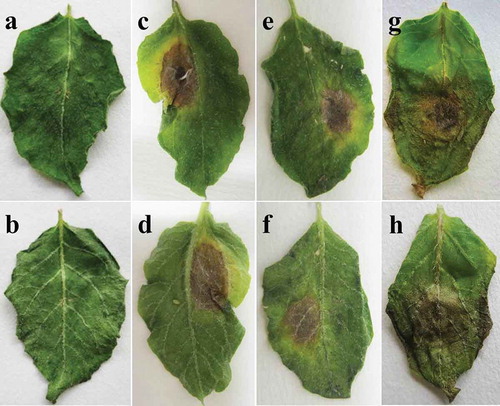
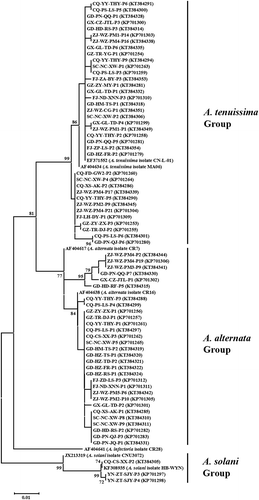
Fig. 3 Phylogenetic tree constructed based on the partial coding sequences of histone 3 gene of 37 A. tenuissima isolates, 30 A. alternata isolates, three A. solani isolates, as well as seven reference sequences retrieved from GenBank. The tree was constructed by neighbour joining using the Kimura two-parameter distance method. Bootstrap values (in percentage) above 70 from 1000 pseudo-replicates are shown for major lineages within the tree.