Figures & data
Table 1. Isolates of Corynespora cassiicola used in the study.
Table 2. Primers used in the iPBS analyses.
Fig. 1 A representative DNA profile of Corynespora cassiicola isolates from different hosts obtained by iPBS analysis using the iPBS primer i2395. Lanes: 1, YPH; 2, YCH; 3, SC1; 4, JD1; 5, DX; 6, KHDL; 7, TTN; 8, JD4; 9, LJ2; 10, JD2; 11, KG1; 12, KG3; 13, KG6; 14, KG8; 15, JD5; 16, JD6; 17, FQ1; 18, FQ2; 19, FQ7; 20, FQ10. M, DNA marker DL5000. The hosts of YPH, YCH, SC1, DX, KHDL, TTN and LJ2 were poinsettia (Euphorbia pulcherrima), scarlet sage (Salvia splendens), lettuce (Lactuca sativa), lilac (Syzygium aromaticum), lipstick (Aeschynanthus pulcher), crane flower (Strelitzia reginae) and hot pepper (Capsicum annuum), respectively. The host of JD1, JD2, JD4, JD5 and JD6 was cowpea (Vigna unguiculata). The host of KG1, KG3, KG6 and KG8 was bitter gourd (Momordica charantia). The host of FQ1, FQ2, FQ7 and FQ10 was tomato (Solanum lycopersicum).
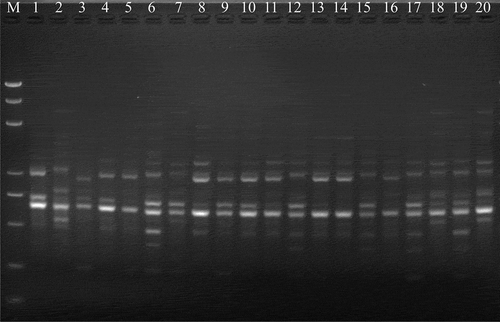
Fig. 2 (Colour online) An unweighted pair group method with arithmetic mean (UPGMA) dendrogram of 69 C. cassiicola isolates obtained from Dicer’s coefficient similarity matrix using iPBS data. The vertical line is represented as a threshold line (the threshold value was 0.564). IG1 and IG2 represent two groups at the threshold value of 0.564.
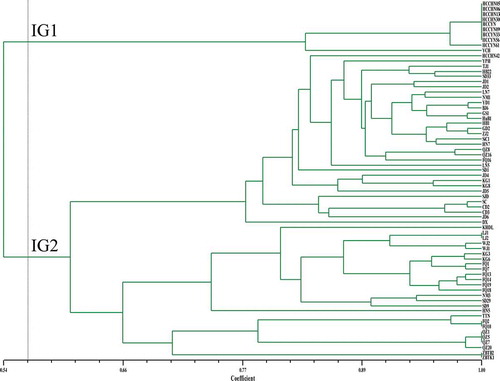
Table 3. Pathogenicity profiles for 44 C. cassiicola isolates.
Fig. 3 (Colour online) The unweighted pair group method with arithmetic mean (UPGMA) dendrogram of 44 C. cassiicola isolates based on the pathogenicity profiles on eight vegetable crops: cucumber, bitter gourd, tomato, eggplant, pepper, bean, cowpea and lettuce. The vertical line is represented as a threshold line (the threshold value was 0.56). PG1 and PG2 represent two groups at a threshold value of 0.56.
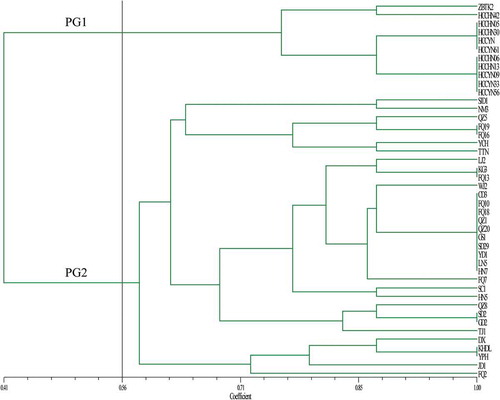
Table 4. Nei’s genetic identity and genetic distance of 16 populations of C. cassiicola detected by iPBS.
Table 5. Genetic diversity of 16 populations of C. cassiicola detected by iPBS.
Table 6. Genetic diversity among 69 isolates of C. cassiicola based on iPBS.
