Figures & data
Table 1. Table of illumina HiSeq 2500 paired end reads obtained and contigs assembled from the Stac-3B and from Stac-4B samples of sweet cherry (Prunus avium) ‘Staccato’.
Table 2. Oligonucleotide primers based on the sequences of fabaviruses infecting sweet cherry (Prunus avium) ‘Staccato’.
Table 3. Pairwise comparisons of the deduced amino acid sequences of the RNA1 segments of the fabaviruses infecting ‘Staccato’ cherry, including also fabavirus sequences in GenBank with Prunus spp. as hosts. The fabavirus-associated sequences detected in ‘Staccato’ cherry are bolded. Sequences determined to be cherry virus F are marked with an asterisk. Numbers below the diagonal show the identities (%) for each comparison and the number of residues that differed for each comparison are shown above the diagonal. The data were obtained using MEGA6 (Tamura et al. Citation2013).
Table 4. Pairwise comparisons of the deduced amino acid sequences of the RNA2 segments of the fabavirus-associated sequences detected in ‘Staccato’ cherry, including also fabavirus sequences in GenBank with Prunus spp. as hosts. The fabavirus-associated sequences detected in ‘Staccato’ cherry are bolded. Sequences determined to be cherry virus F are marked with an asterisk. Numbers below the diagonal show the identities (%) for each comparison and the number of residues that differed for each comparison are shown above the diagonal. The data was obtained using MEGA6 (Tamura et al. Citation2013).
Fig. 2 (Colour online) Schematic 3diagram of the genome organization of the proposed and RACE-confirmed Prunus virus F (PrVF) isolate CP1 (Stac-3B_C3 plus Stac-3B_c11) and cherry virus F (CVF) isolate CC1 (Stac-3B_C4 plus Stac-3B_c7). RNA1 and RNA2 segments are shown in blue and green, respectively, with CVF in darker shades than PrVF. Untranslated regions (UTRs) are shown in yellow. Vertical lines demarcating gene products in the coding sequences of each segment are placed at the protease cleavage sites. Abbreviations are as follows: NTP-BPP = nucleoside triphosphate binding protein peptide; RdRp = RNA dependent RNA polymerase; MP = movement protein; CP(L) = large capsid protein; CP(S) = small capsid protein. The putative genome-linked protein (VPg) is located between NTP-BPP and 2protease and is marked with an asterisk. Figure layout is adapted from Koloniuk et al. (Citation2018).
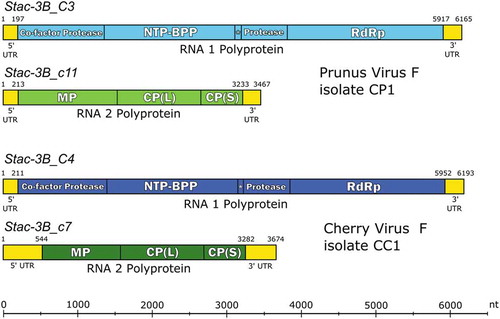
Table 5. Deduced proteins, predicted sizes and associated protease cleavage sites of Prunus virus F (PrVF) putative isolate CP1 (Stac-3B_C3/_c11). The corresponding values for cherry virus F (CVF) putative isolate CC1 (Stac-3B_C4/_c7) are indicated in parentheses, except in the case of the cleavage sites that are shown in separate columns. The unique CP(L)/CP(S) dipeptide cleavage site for CVF-CC1 is bolded.
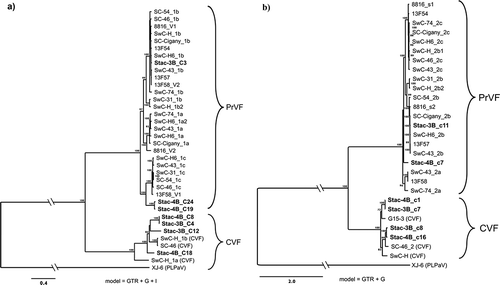
Fig. 1 Bayesian phylogenetic trees illustrating the relationships between the isolates of fabaviruses detected in sweet cherry cv. Stacatto in Canada and published sequences of fabaviruses infecting Prunus spp. Trees based on available RNA1 genome sequences (a) RNA2 genome sequences (b), deduced amino acid sequences (aa) of the Pro-Pol region (c) and the deduced aa sequences of the CP (S + L) region (d) are shown. Posterior probabilities of 70% or higher are shown at group nodes. Scale bars indicate the number of substitutions per residue.
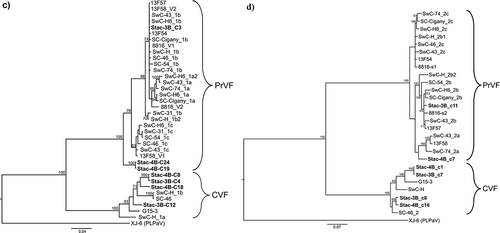
Fig. 3 (Colour online) Predicted secondary structures for: a highly conserved 28 nucleotide (nt) region at the proximal end of the 5ʹUTR (a) and a 106 nt region that is conserved among the 3ʹUTR (b) of the RNA1 and RNA2 of the putative Canadian virus isolates PrVF-CP1 and CVF-CC1 that were RACE-confirmed. Variable nucleotide positions are identified where a single bar above and a double bar below indicate that the mutation occurs in CVF RNA1 and CVF RNA2, respectively. An asterisk indicates a wobble G-U pairing that may not occur in all isolates considered and the nt in parentheses indicates a mutation identified in PrVF-CP1 RNA2.
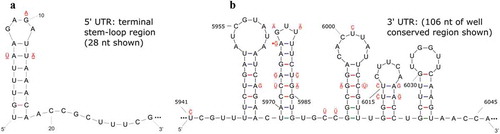
Fig. 4 (Colour online) Schematic diagram showing the variability that was identified in the 3ʹUTR of RNA2 for cherry virus F (CVF) and Prunus virus F (PrVF). The location of partial CP coding sequence and the conserved region of the 3ʹ UTR are shown in green and yellow, respectively. Graduated crosshatching is included to illustrate the nature of repeats found in the sequences of various PrVF isolates, focussing on PrVF isolate 8816_s2. The upstream variable region of the 3ʹ UTR is shown in orange. The large downstream indels are shown in purple, labelled INDEL (392 nt) and INDE (truncated, 318 nt). Short olive-coloured sections are associated with the repeat sequence and do not seem to have a counterpart in the prevailing default pattern. The two locations in the 3ʹUTR at which the known ‘non-default’ features occur are indicated with dashed lines. At both of these locations, multiple variants have been seen. These are indicated with bold curved brackets, and representative isolates of both CVF and PrVF bearing these features are listed as examples.
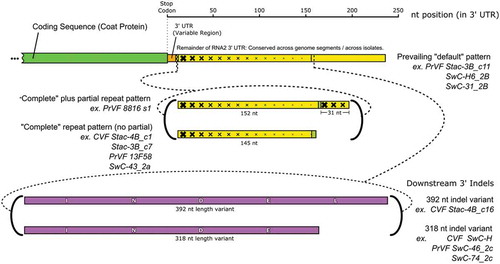
Table 6. RDP4 results following the analysis of Prunus virus F (PrVF) isolate SwC 74_2c indicating that it is a recombinant with PrVF 8816_s1 as the major parent and cherry virus F isolate SwC_H as the minor parent. This was supported by all seven methods used for screening. The P values obtained and the references for the individual methods are provided.
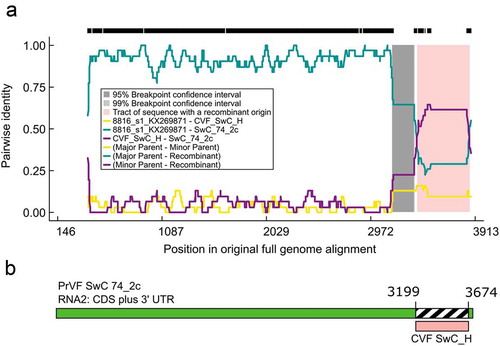
Fig. 5 (Colour online) Schematic representation of a putative recombination event detected in some Prunus virus F (PrVF) isolates with SwC 74_2c as a representative and showing PrVF 8816_s1 as the major parent and cherry virus F (CVF) isolate SwC_H as the minor parent, illustrated using an RDP Plot (a) and a schematic of the SwC 74_2c genome showing the recombination event at the 3ʹ end of the virus (b). The 5ʹ termini were excluded from the multiple sequence alignment for ease of RDP4 analysis. The numbers on the RDP Plot and the genome map indicate nucleotide positions (nt) and the putative nt positions of recombination breakpoints, respectively.