Figures & data
Table 1. Potyvirus infection of cultivated legume samples collected from different provinces surveyed in Iran1.
Table 2. List of primers used in reverse transcription-polymerase chain reaction (RT-PCR) tests and sequencing.
Table 3. List of virus isolates studied.
Fig. 1 Reverse transcription-polymerase chain reaction (RT-PCR) using universal primers () designed to amplify regions in potyvirus genomes. (A) Amplification of the expected DNA fragments (~300 bp) using NIb primers (lanes 1–4 and 6); (B) Specific and non-specific amplifications of DNA amplicons of ~300 bp (lane 3) and 500 bp (lane 1), respectively, using NIb primers; (C) Amplification of the expected DNA fragments (~700 bp) using WCIEN primers (lanes 2–7); and (D) Amplification of the expected DNA fragments (~700 bp) using NWCIEN primers (lanes 1–6). GeneRuler 100 bp DNA ladder (Thermo Scientific, MA, USA) was used in gel electrophoresis. The PCR products and the DNA ladder were visualized using GelRed (Biotium Inc., CA, USA) staining.
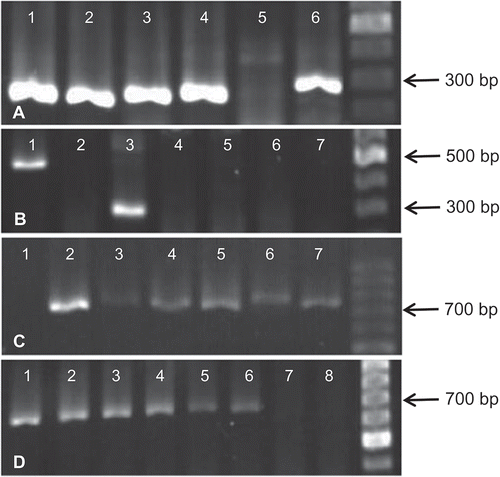
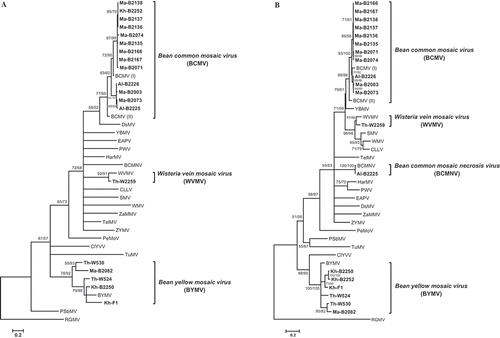
Fig. 2 A maximum likelihood (ML) tree constructed from: (A) the partial nucleotide sequence of the nuclear inclusion b (NIb) region, and (B) coat protein (CP) region for 19 Iranian virus isolates from legumes (shown in bold; ) and 20 representative Potyvirus species listed in Supplementary Table 1. Numbers at each node indicate the percentage of supporting bootstrap samples in ML and NJ methods, respectively. The homologous sequence of an isolate of Ryegrass mosaic virus (RGMV; genus Rymovirus) was used as the outgroup.