Figures & data
Fig. 1 (Colour online) Symptoms observed in pepper open fields in Comarca Lagunera (CL). (a) Map of CL which includes surveyed counties of Durango state (blue) and Coahuila state (green) of CL. (b) A pepper plant showing yellow mosaic, deforming and chlorotic leaf. (c) A stunted pepper plant with severe leaf deformation and yellowing. (d) A pepper plant with chlorotic leaf and yellow mosaic. (e) General view of pepper open field surveyed in 2016 with a 90% disease incidence caused by begomoviruses when double or triple infections were detected.
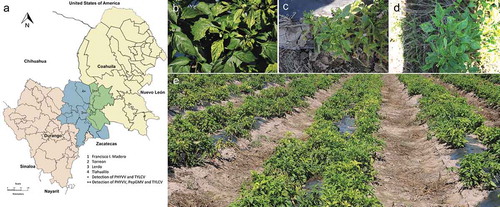
Fig. 2 Phylogenetic trees based on multiple sequence alignment of the complete genome components. (a) DNA-A. (b) DNA-B. Phylogenetic trees were constructed using maximum likelihood method. Numbers represent bootstrap percentages values out of 1000 replicates using MEGA 7. Nodes with credibility values of 65% or high are shown. Beet curly top virus (BCTV) was used as a root. Sequences of begomoviruses isolated in this work are underlined. Acronym of virus sequences were as follows: Pepper golden mosaic virus (PepGMV), Pepper huasteco yellow vein virus (PHYVV), Tomato yellow leaf curl virus (TYLCV), Rynchosia golden mosaic virus (RhGMV), Chino del tomate virus (CdTV), Sida mosaic Sinaloa virus (SiMSV), Tomato severe leaf curl virus (ToSLCV), Tomato chino La Paz virus (ToChLPV), Abutilon golden mosaic Yucatan virus (AbGMYV), Euphorbia mosaic virus (EuMYV), Tomato golden mottle virus (ToGMV). The abbreviations after the virus acronyms represent the states of Mexico where the isolates were obtained: SIN (Sinaloa), CHI (Chiapas), YUC (Yucatan), SLP (San Luis Potosí), BC (Baja California), SON (Sonora), QTO (Querétaro), TAM (Tamaulipas), MX (No knowledge of the state of collection) and CL (La Comarca Lagunera). GenBank accession numbers are shown beside the names of each isolate.
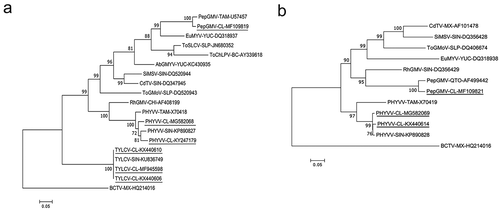
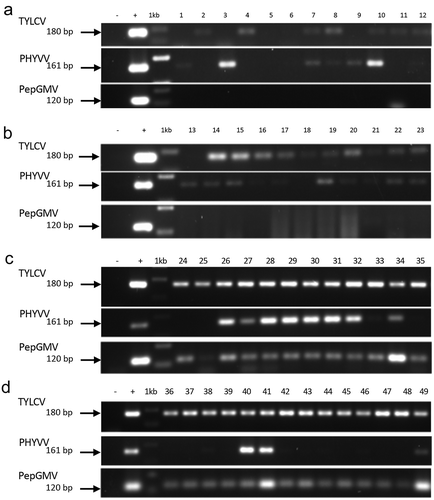
Fig. 3 Detection by PCR using specific primers for the begomoviruses TYLCV, PHYVV and PepGMV. (a) Samples collected in 2014 in CL open fields at Torreón, Coahuila. (b) Samples collected in 2015 in CL open fields at Tlahualilo, Durango. (c and d) Samples collected 2016 in CL open fields at Francisco I. Madero, Coahuila. Negative control (−), DNA of PHYVV (+) as positive control, 1Kb molecular maker (Invitrogen, USA).