Figures & data
Fig. 1. Eggplants showing phytoplasma symptoms. (a), comparison between symptomatic and asymptomatic plants; (b), general symptoms on leaves showing phyllody and witches’ broom; (c), leaves showing witches’ broom; (d), flowers showing phyllody.

Fig. 2. Phylogenetic tree constructed from 16S rDNA sequence data by maximum likelihood. Thirty-eight 16S rDNA sequences deposited in GenBank were used, with Acholeplasma laidlawii used as the outgroup. The numbers on the branches are the bootstrap support obtained from 1000 replicates. The red star indicates the location of China EPP within the phylogenetic tree. BLL, brinjal little leaf. EBB, eggplant big bud. EPP, eggplant phyllody phytoplasma. EGC, eggplant giant calyx.
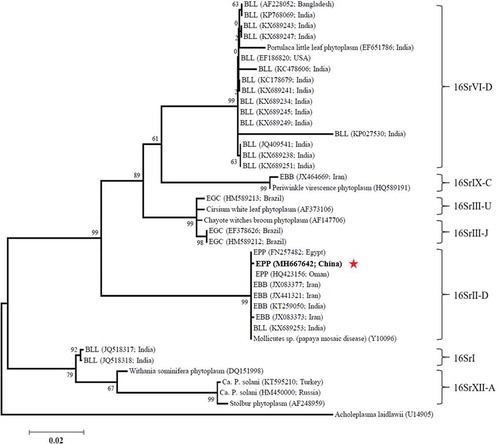
Fig. 3. Phylogenetic tree constructed from secA gene sequence data by maximum likelihood. Thirty-six secA sequences deposited in GenBank were used, with Bacillus subtilis W168 used as the outgroup. The numbers on the branches are the bootstrap support obtained from 1000 replicates. The red star indicates the location of China EPP within the phylogenetic tree.
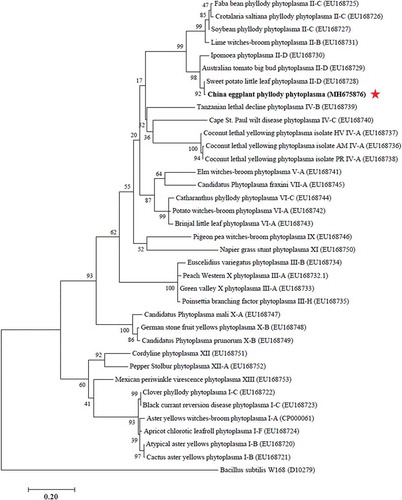
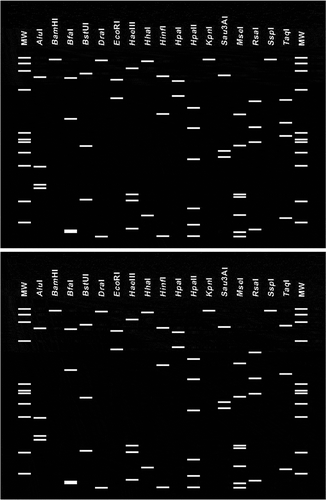
Fig. 4. Virtual RFLP patterns from in silico digestion of the 16S rDNA sequence. The 16S rDNA sequence from Candidatus Phytoplasma australasia (GenBank Y10096) (bottom) was used as the reference strain for the 16SrII-D subgroup. Simulated digestions were performed with 17 endonucleases: AluI, BamHI, BfaI, BstUI (ThaI), DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, MboI, MseI, RsaI, SspI and TaqI. MW indicates the PhiX174DNA-HaeIII marker.