Figures & data
Table 1. Phytoplasma gene sequences used in this study
Fig. 1 Phylogenetic tree of the 16S rRNA gene sequences from the 21 published phytoplasma strains using (a) Neighbour Joining method (NJ) and (b) Maximum Likelihood method (ML); the trees were rooted using Bacillus subtilis (AB042061)
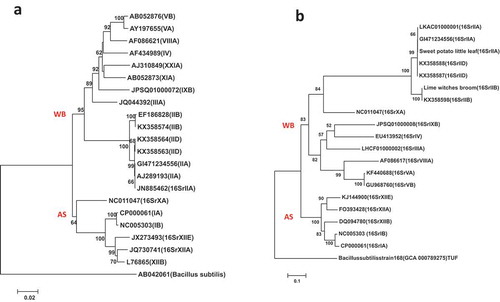
Fig. 2 Phylogenetic trees of tuf gene sequences from the previously published 19 phytoplasma strains () using the NJ method; (a) shows the tree based on the short DNA sequence (about 350 bp), (b) shows the tree based on the long DNA sequence (about 730 bp). The trees were rooted using Bacillus subtilis strain 168 (GCA:000789275)
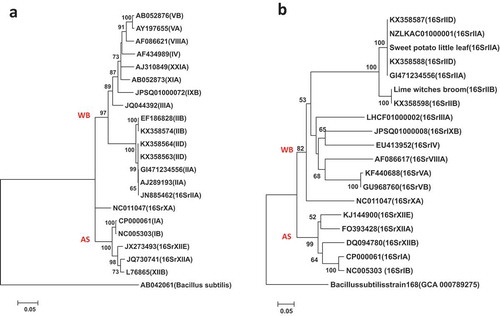
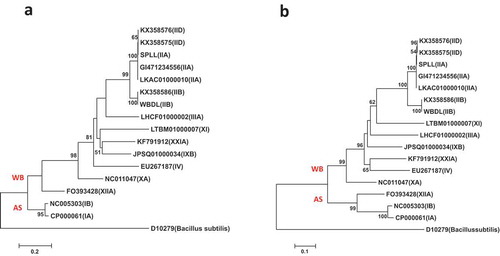
Fig. 3 Phylogenetic trees of secA gene sequences from the previously published 16 phytoplasma strains () using the NJ method; (a) shows the tree based on the short DNA sequence (about 530 bp), (b) shows the tree based on the long DNA sequence (about 1220 bp). The trees were rooted using Bacillus subtilis (D10279)
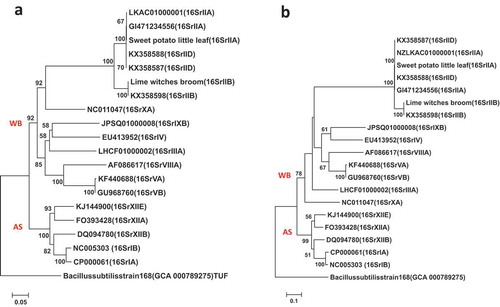
Fig. 4 Phylogenetic trees of tuf gene sequences from the previously published 19 phytoplasma strains () using the ML method; (a) shows the tree based on the short DNA sequence (about 350 bp), (b) shows the tree based on the long DNA sequence (about 730 bp). The trees were rooted using Bacillus subtilis strain 168 (GCA:000789275)
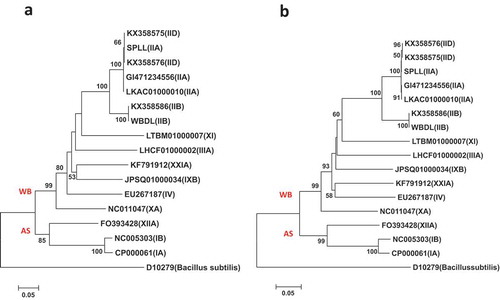
Fig. 5 Phylogenetic trees of secA gene sequences from the previously published 16 phytoplasma strains () using the ML method; (a) shows the tree based on the short DNA sequence (about 530 bp), (b) shows the tree based on the long DNA sequence (about 1220 bp). The trees were rooted using Bacillus subtilis (D10279)