Figures & data
Fig. 1 Symptoms of leaf yellowing (a, b), reduction in internodes (c), leaf stunting (d), and dryness and collapse of older fronds (e) in Roystonea regia.
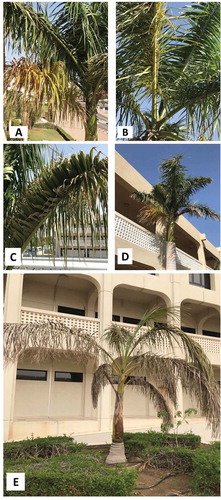
Table 1. The 16S rDNA sequences of different phytoplasma strains obtained from the National Centre for Biotechnology Information (NCBI) GenBank database for phylogenetic analysis
Table 2. Representative phytoplasma 16S rRNA, tuf, secA, and imp gene sequences from the National Centre for Biotechnology Information (NCBI) GenBank database used in phylogenetic analysis
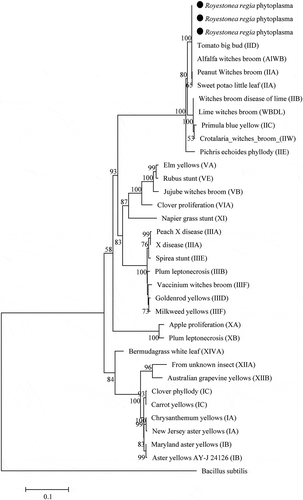
Fig. 2 Phylogenetic tree based on the 16S ribosomal RNA gene sequences of Roystonea regia phytoplasmas (black circles) with 41 phytoplasma strains from different groups and subgroups. The tree was rooted using Bacillus subtilis (AB04261). The phylogenetic tree was constructed by the Maximum Likelihood method and units are the number of base substitutions per site. Bootstrap values are expressed as percentage of 1,000 replicates
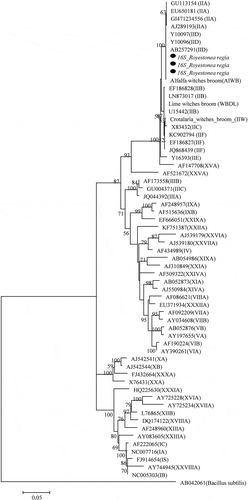
Fig. 3 Phylogenetic tree based on tuf gene sequences of Roystonea regia phytoplasmas (black circles) with 41 phytoplasma strains from different groups and subgroups. The tree was rooted using Bacillus subtilis (GCA000789275). The phylogenetic tree was constructed by the Maximum Likelihood method and units are the number of base substitutions per site. Bootstrap values are expressed as percentage of 1,000 replicates
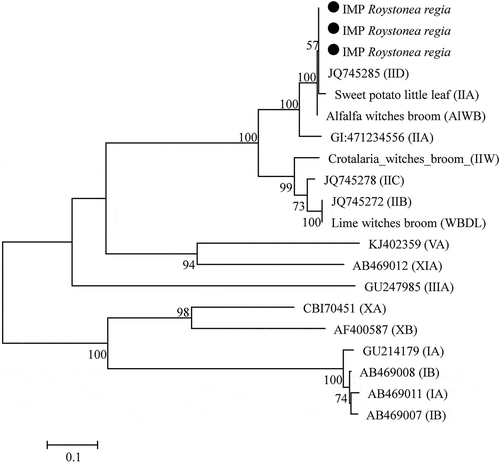
Fig. 4 Phylogenetic tree based on imp gene sequences of Roystonea regia phytoplasmas (black circles) with 17 phytoplasma strains from different groups and subgroups. The phylogenetic tree was constructed by the Maximum Likelihood method and units are the number of base substitutions per site. Bootstrap values are expressed as percentage of 1,000 replicates
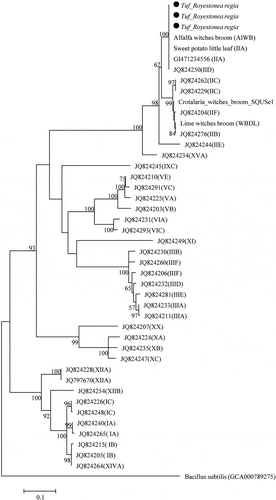
Fig. 5 Phylogenetic tree based on the combined 16S ribosomal RNA, tuf and imp gene sequences of Roystonea regia phytoplasmas (black circles) with 41 phytoplasma strains from different groups and subgroups. The tree was rooted using Bacillus subtilis (GCA000789275). The phylogenetic tree was constructed by the Maximum Likelihood method and units are the number of base substitutions per site. Bootstrap values are expressed as percentage of 1,000 replicates