Figures & data
Table 1. Strains of Pseudopyrenochaeta lycopersici examined in this study
Table 2. Primers used in this study
Fig. 1 Alignment of deduced amino acid sequences of the (a) alpha box, (b) HMG box and amino acid identity and similarity to Pseudopyrenochaeta lycopersici (Pl) of Ascochyta lentis (Al), A. rabiei (Ar), Alternaria alternata (Aa), Cochliobolus heterostrophus (Ch), Leptosphaeria maculans (Lm), Phaeosphaeria nodorum (Pn), Neurospora crassa (Nc), Podospora anserina (Pa) and Pyrenopeziza brassicae (Pb). Amino acid residues shaded in black are identical, while those in grey are similar. The primers Sn-ab1, Sn-ab2, Pl-alpha-2, Sn-HMG1 and Sn-HMG2 are indicated as dotted lines. The positions of introns are shown as vertical arrows
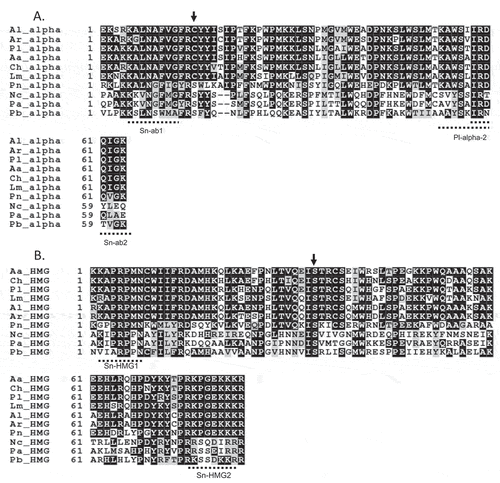
Fig. 2 Alignment of the deduced amino acid sequence of ORF1 genes in MAT1-1 and MAT1-2 isolates of Cochliobolus heterostrophus (Ch) GenBank accession nos. AF029913 (MAT1-1) and AF027687 (MAT1-2), Phaeosphaeria nodorum (Pn) GenBank accession nos. AY212018 (MAT1-1) and AY212019 (MAT1-2), Leptosphaeria maculans (Lm) GenBank accession nos. AY174048 (MAT1-1) and AY174049 (MAT1-2) and Pseudopyrenochaeta lycopersici (Pl). The arrow indicates the beginning of idiomorph
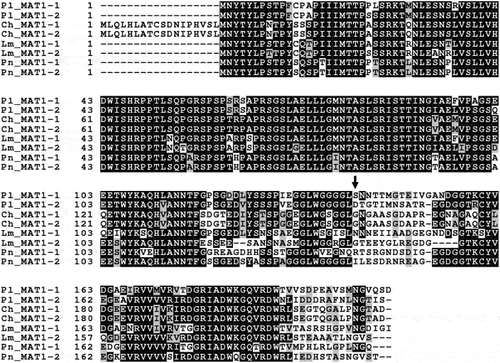
Fig. 3 Dot-plot comparison of MAT loci. The MAT1-1 idiomorph starts from 355 bp and ends at 3295 bp. The MAT1-2 idiomorph starts from 133 bp and ends at 3551 bp. Each ORF is indicated as arrows
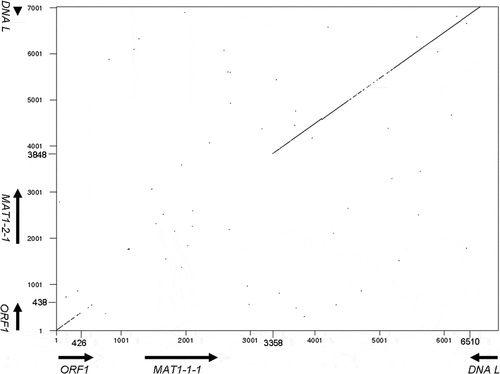
Fig. 4 PCR cloning of MAT loci from Pseudopyrenochaeta lycopersici type 1. (a) Amplification of the sequence corresponding to the alpha box using degenerate primers 1, 2 and 3 from strain CBS267.59 of the fungus. (b) TAIL-PCR to obtain the 5ʹ and 3ʹ region sequence from the alpha box. (c) RT-PCR with primers 6 and 7, 3ʹ-RACE with primers 10, 11, 12 and 13 and multiplex PCR with primers 6 and 7. (d) Amplification of the sequence corresponding to the HMG box from strain Oha3-6 using degenerate primers 4 and 5. (e) Amplification of the MAT1-2 idiomorph using primers 25, 26, 27 and 28 designed based on the sequence of the HMG box and primers 29, 30 and 31 designed based on the sequences of DNA lyase and ORF1 gene. (f) RT-PCR with primers 8 and 9, 3ʹ-RACE with primers 8, 10, 11 and 14 and multiplex PCR with primers 8 and 9. (g) Structural organization of the P. lycopersici type 1 MAT loci. Grey boxes indicate MAT idiomorphs. Scale bar is marked to denote the entire region sequenced. Direction of transcription and limits of the ORFs is indicated by arrows, with a horizontally striped and a vertically striped box representing the alpha and HMG boxes in MAT1-1-1 and MAT1-2-1, respectively. 3ʹ-UTR are shown as dotted lines. Introns are indicated by diamonds on the striped boxes. Primers are indicated as arrowheads
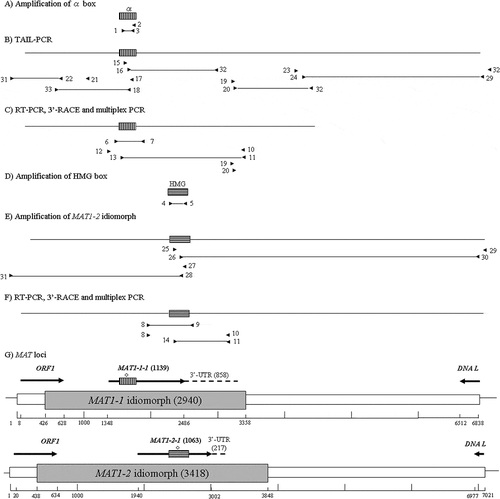
Fig. 5 Expression of the mating type genes in Pseudopyrenochaeta lycopersici. gDNA, genomic DNA of strain CBS267.59 (MAT1-1) and Oha3-6 (MAT1-2) as the template. + = RT-PCR with reverse transcriptase; – = without reverse transcriptase. Genomic DNA was extracted from each strain grown on PDB (potato dextrose broth). Total RNA was extracted from each strain grown on PDB or CD (Czapek-dox)
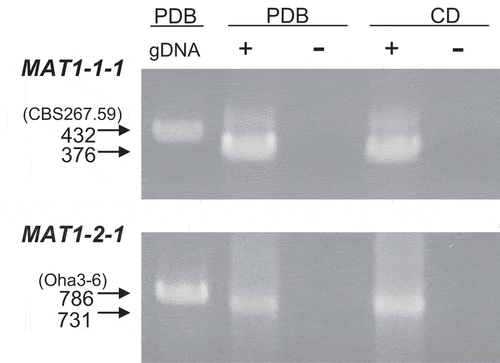
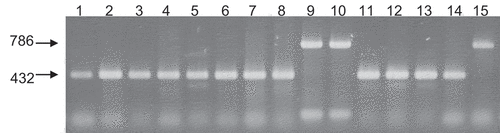
Fig. 6 Multiplex PCR with primers 6, 7, 8 and 9. From left to light: lane 1, CBS267.59; lane 2, MAFF712039; lane 3, Au3-10; lane 4, Au4-3; lane 5, Ty2-7; lane 6, Lt2-12; lane 7, GR2-1; lane 8, GR3-3; lane 9, Y502-H1; lane 10, Y502-M33; lane 11, Y519-31; lane 12, 24-C; lane 13, PrF2; lane 14, Oha3-5; lane 15, Oha3-6. Lanes 1–8 and 11–14 are MAT1-1 and lanes 9, 10 and 15 are MAT1-2.