Figures & data
Fig. 1 Counties and municipal districts from which Plasmodiophora brassicae-infected canola root galls were collected and subjected to molecular analysis. Sampled counties and municipal districts are highlighted in grey, with the number of samples indicated in parentheses. The geographic shape file for Canada is from Boundary Files, 2011 Census. Statistics Canada Catalogue no. 92–160-X. Reproduced and distributed on an ‘as is’ basis with the permission of Statistics Canada. The Alberta municipal boundaries shape file is from AltaLIS and is used under the Open Data Licence.
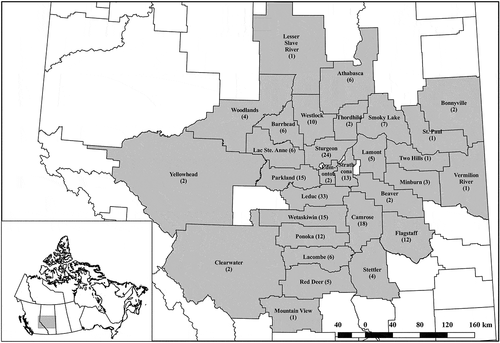
Table 1. Clubbed canola roots infected predominantly with ‘population two’ of Plasmodiophora brassicae. Percentage of P. brassicae DNA that was determined to be ‘population two’ for each assay is shown, as determined by comparison with quantities determined for the ‘population one’ P. brassicae assays Pb_pop1_52 and Pb_pop1_255
Fig. 2 (Colour online) Maps of central Alberta showing the detection of the original ‘population one’ and ‘population two’ of Plasmodiophora brassicae in canola root galls over time. (A) Samples collected from 2005 to 2007. (B) Samples collected from 2008 to 2010. (C) Samples collected from 2011 to 2013. (D) Samples collected from 2014 to 2016. The P. brassicae strains were determined by qPCR analysis. Green circles indicate only ‘population one’ strains were detected. Yellow triangles indicate trace levels of ‘population two’ were detected (<1% of total P. brassicae DNA). Red stars indicate galls predominantly infected (>99%) by ‘population two’ strains. The Alberta municipal boundaries shape file is from AltaLIS and is used under the Open Data Licence.
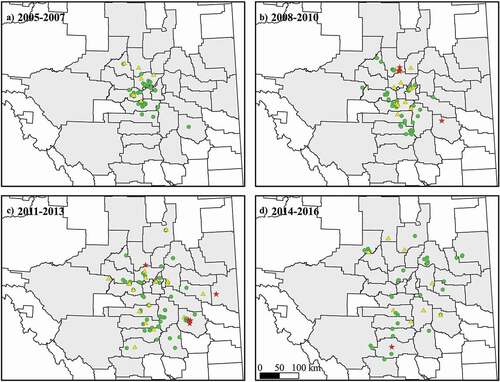
Table 2. Correlations between quantities of DNA determined by the molecular assays used on Plasmodiophora brassicae-infected canola roots
