Figures & data
Fig. 1 Location (counties) in Minnesota where soybean stems were collected and tested for the presence of Diaporthe spp. (shaded). Symbols indicate where Diaporthe caulivora (●), D. longicolla (X), and D. cucurbitae (▲) were detected. Includes plants with symptoms of pod and stem blight, stem canker, and top dieback, as well as asymptomatic plants.
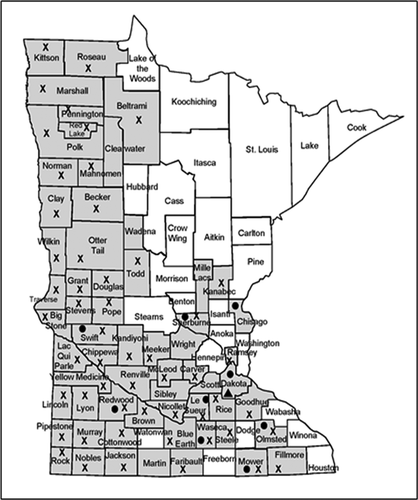
Table 1. Isolate code, species, geographical origin, associated symptoms, source, and virulence group for isolates of Diaporthe spp. used in these studies.
Fig. 2 Phylogenetic analysis based on combined ITS and TUB DNA sequences. The tree is drawn to scale with branch lengths measured in the number of substitutions per site. Bootstrap values with 1000 replications are shown at the nodes. Isolate codes noted as DL are Diaporthe longicolla and DC are D. caulivora. The D. longicolla, D. caulivora, and D. cucurbitae sequences are from isolates collected in this study and the others are from GenBank for comparison ().
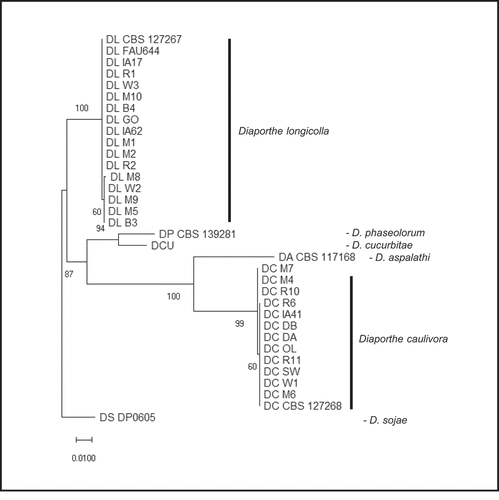
Table 2. Diaporthe spp. detected in soybean stems with different types of symptoms.
Table 3. Virulence of 11 isolates of Diaporthe caulivora (Dc) and 13 isolates of D. longicolla (Dl) on six soybean cultivars. Virulence was measured as percentage of internode that was symptomatic 6 weeks post-inoculation in a greenhouse at 24°C. Treatments in the same column not designated by the same letter are significantly different (α = 0.05).
Fig. 3 Average virulence of 11 isolates of D. caulivora (cross hatch bars) and 13 isolates of Diaporthe longicolla (black bars) on six soybean cultivars. Virulence was measured as percentage of internode length with brown lesions 6 weeks after inoculation in a greenhouse at 24°C. Each cv. was analysed separately. Treatments designated by different letters are significantly different (α = 0.05).
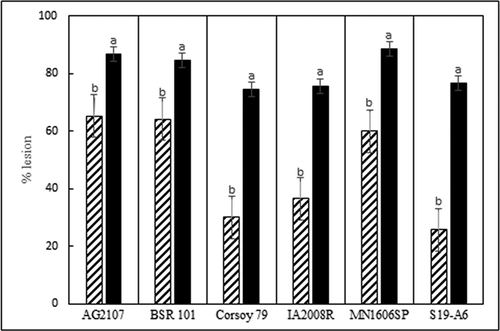
Fig. 4 Branch dieback (average) resulting from inoculation of six soybean cultivars with 11 isolates of Diaporthe caulivora (cross hatch bars) and 13 isolates of D. longicolla (black bars), 6 weeks post inoculation in a greenhouse at 24°C. Percentage branch dieback was determined as the number of dead branches/total number of branches on each plant x 100. Each cv. was analysed separately. Treatments noted with different letters are significantly different (α = 0.05).
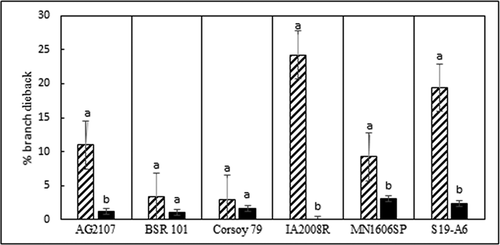
Fig. 5 Average colony diameter (cm) of Diaporthe caulivora (cross hatch bars) and D. longicolla (black bars) isolates after 14 d of growth on 0.5x PDA at three temperatures. Each temperature was analysed separately. Treatments designated by different letters are significantly different (α = 0.05).
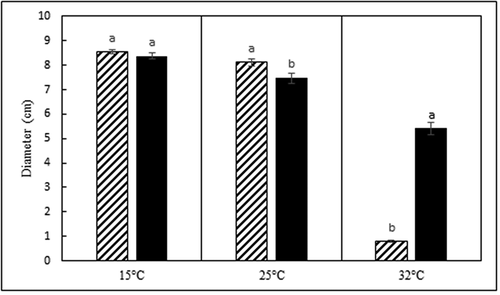
Table 4. Sensitivity of 11 isolates of Diaporthe caulivora (Dc) and 13 isolates of D. longicolla (Dl) to the fungicides pyraclostrobin and tebuconazole. The EC50 (µg mL−1 a.i.) was determined based on radial growth on agar plates containing these fungicides. Fungicides were analysed separately. Treatments in the same column not designated by the same letter are significantly different (α = 0.05).
