Figures & data
Table 1. Claviceps spp. described between 1853 and 1940s.
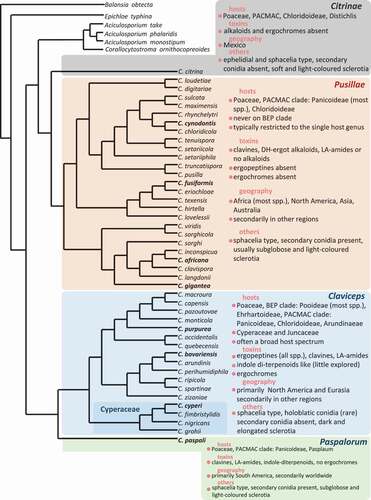
Fig. 1 (Colour online) Phylogenetic relationships of ergot species with available DNA sequences. The classification into four sections (Citrinae, Claviceps, Paspalorum, Pusillae) and their characterization (toxicity, host range, geography and others), were based on.Píchová et al. (Citation2018) and Liu et al. (Citation2020). Ergot species in bold are of agricultural importance: C. africana on sorghum, C. cynodontis on Bermuda grass, C. fusiformis on pearl millet, C. gigantea on maize, C. paspali on paspalum and C. purpurea on cereals and forage grasses are important food and feed contaminants causing toxicity to human and livestock. Claviceps bavariensis infecting common pasture grasses may impact livestock. The maximum likelihood tree generated in PhyML 3.0. was constructed using ITS-LSU rDNA, TUB2, TEF1-α and RPB2 sequences from Píchová et al. (Citation2018) and Liu et al. (Citation2020).