Figures & data
Table 1. Virulence phenotypes and group designations of Puccinia triticina isolates
Fig. 1. Distributions of 14 classes of di- and tri-nucleotide simple sequence repeat motifs in the EST database of P. triticina. Classes of simple sequence repeats described by Jurka & Pethiyagoda (Citation1995) and Katti et al. (Citation2001) were listed as following: 1 = AT/AT; 2 = AG/GA/CT/TC; 3 = AC/CA/TG/GT; 4 = GC/CG; 5 = AAT/ATA/ TAA/ATT/TTA/TAT; 6 = AAG/AGA/GAA/CTT/TTC/TCT; 7 = A AC/ACA/CAA/GTT/TTG/TGT; 8 = ATG/TGA/GAT/CAT/A TC/TCA; 9 = AGT/GTA/TAG/ACT/CTA/TAC; 10 = AGG/GGA/GAG/ CCT/CTC/TCC; 11 = AGC/GCA/CAG/GCT/CTG/TGC; 12 = ACG/ CGA/GAC/CGT/GTC/TCG; 13 = ACC/CCA/CAC/GGT/GTG/TGG; 14 = GGC/GCG/CGG/GCC/CCG/CGC.
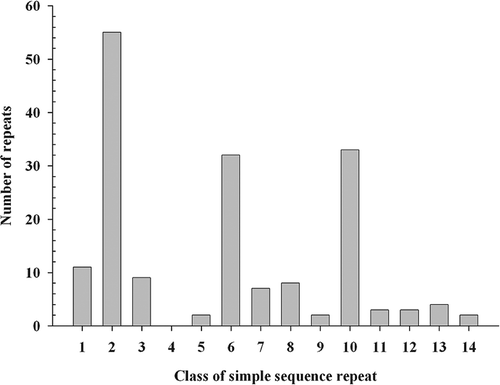
Table 2. Repeat motif, primer sequence, amplification conditions and characteristics
Fig. 2. The clustering structure in the data sets derived from virulence and EST-SSRs. a, Functions of K with the number of clusters for Puccinia triticina isolates based on virulence (□) and EST-SSR genotypes (▪) and b, two-dimensional depiction of clusters at K = 12 differentiated by virulence to 16 differential lines (□, ▵, ∘) and 21 EST-SSR markers (▪, ▴, •). Isolates were grouped based on the virulence to resistance gene Lr2a, Lr2c and Lr17a. Group I (□,▪) are avirulent to Lr2a and Lr2c but virulent to Lr17a. Group II (▵,▴) are avirulent to Lr2a, Lr2c and Lr17a. Group III (∘, •) are virulent to Lr2a and Lr2c but avirulent to Lr17a.
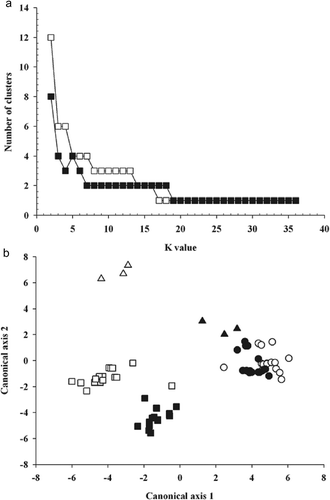
Fig. 3. Genetic dissimilarity dendrogram of 35 Puccinia triticina isolates collected in Canada based on the unweighted pair group method with arithmetic means clustering method using 1-Dice coefficient calculated from 21 EST-SSR markers. Numbers along the nodes are bootstrap values > 70% in 1000 replicas. The first four letters of isolate designation represent virulence phenotypes of P. triticina isolates based on 16 differential lines and the last letter represents the location where the isolate was collected. O, isolates collected in Ontario and Quebec; M, isolates collected in Manitoba and Saskatchewan; B, isolates collected in British Columbia and Alberta; P, isolates collected in Prince of Edward Island. Group I, II and III represent three groups of isolates clustered based on virulence to Lr2a, Lr2c and Lr17a.
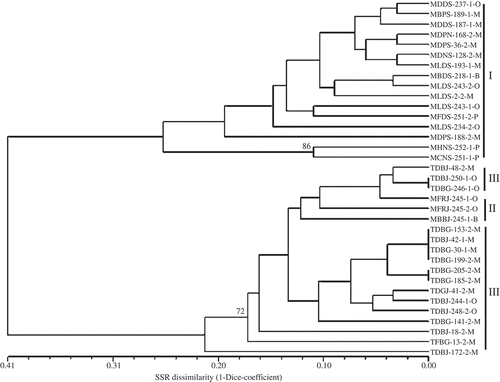
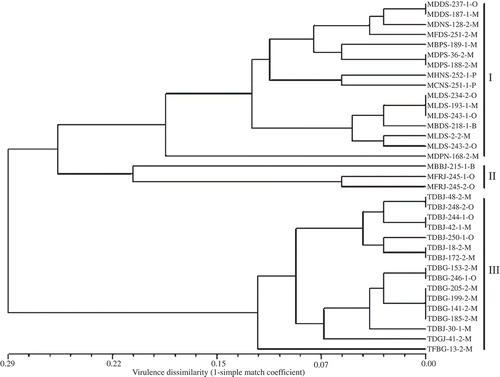
Fig. 4. Virulence dissimilarity dendrogram of 35 Puccinia triticina isolates collected in Canada in 2007 based on the unweighted pair group method with arithmetic means clustering method using simple mismatch coefficient (1-simple match coefficient) calculated based on virulence on 16 leaf rust resistance genes. Numbers along the nodes are bootstrap values > 70% in 1000 replicas. The first four letters of isolate designation represent virulence phenotypes of P. triticina isolates based on 16 differential lines and the last letter represents the location where the isolate was collected. O, isolates collected in Ontario and Quebec; M, isolates collected in Manitoba and Saskatchewan; B, isolates collected in British Columbia and Alberta; P, isolates collected in Prince of Edward Island. Group I, II and III represent three groups of isolates clustered based on virulence to Lr2a, Lr2c and Lr17a.