Figures & data
Figure 1. System-level characterization of metabolic regulation (A) by multi-omics analysis of plant species, natural accessions, mutants, conditions, developmental stages, organs, tissues and organelles (B). Progress in chromatography, mass spectrometry, robotization, sequencing (among others) (C) has enabled the identification and quantification of genomes, transcriptomes, proteomes, and metabolomes (D). Computational analysis of the obtained data led to identifying novel interactions, improved annotation of metabolic pathways, and understanding of signaling and regulatory cascades upstream and downstream of metabolism (E). Figure was created using Biorender.com.
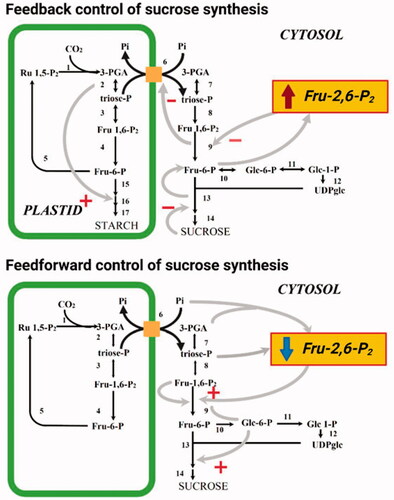
Figure 2. The role of Fru-2,6-P2 in feedback and feedforward control of sucrose synthesis. + represents allosteric activation, - represents allosteric inhibition. Reactions shown are catalyzed by the following enzymes (note in some instances multiple reactions are represented by a single arrow): 1, Rubisco; 2, chloroplastic PGK and chloroplastic TPI; 3, chloroplastic Fru-1-6-P2 aldolase; 4, chloroplastic FBPase; 5, transketolase, sedoheptolase-1,7-bisphosphatase aldolase, sedoheptolase-1,7-bisphosphatase, phosphopentoepimerase, phosphoriboisomerase and phosphoribulokinase; 6, triose phosphate transporter; 7, cytoslic PGK and cytosolic TPI; 8, cytosolic Fru-1-6-P2 aldolase; 9, cytosolic FBPase; 10, cytosolic PGI; 11, cytosolic PGlcM, 12 UGPase, 13 SPS, 14 sucrose phosphatase; 15, choroplastic PGI and chloroplstic PGlcM; 16, AGPase; 17, starch synthase and branching enzyme.