Figures & data
Figure 2. 3D schematic view of the inactive form of D2R in complex with risperidone, which was embedded in the membrane bilayer. Water and ion atoms were rendered by solvation surface and vdW models of VMD, respectively.
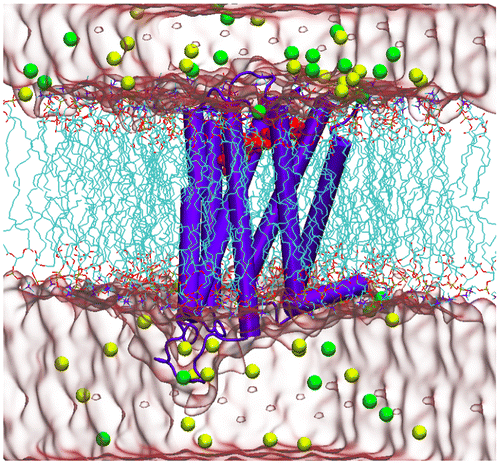
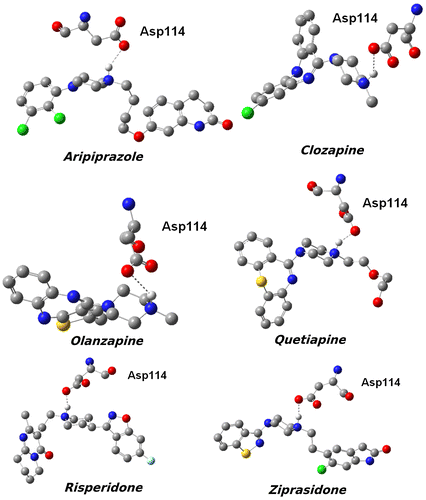
Figure 3. 3D structures of the compounds and their surfaces at the binding site of D2R. The amino acids interacting with the drugs are shown.
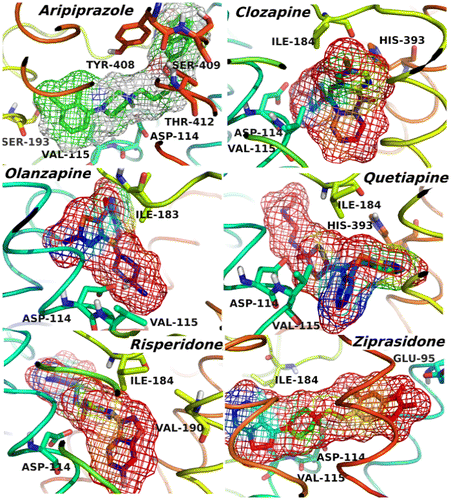
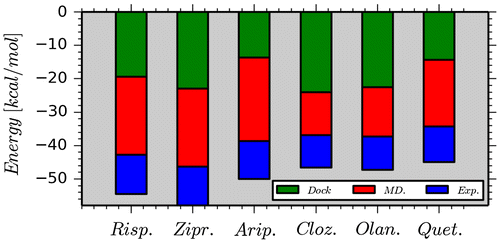
Table 1. Comparison of calculated ligand binding energies with the experimental results.
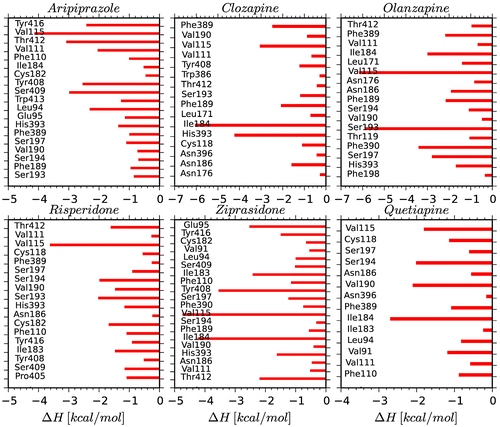
Figure 4. Calculated interaction energies between the drugs and Asp114 using representative structures obtained from the docking and MD simulations. The experimental binding affinities between the drugs and D2R are also given to show more clearly the role of Asp114 in the active site.