Figures & data
Figure 2. Amino acids color-coded (red hydrophobic; blue hydrophilic) based on the values of hydrophobicity scales of: (a) Cornette et al. (Citation1987) (−3.1 to 5.7); (b) Eisenberg et al. (Citation1984) (−2.53 to 1.38); (c) Engelman et al. (Citation1986) (−12.3 to 3.7); (d) Hopp and Woods (Citation1983) (−3.4 to 3.0); (e) Janin (Citation1979) (−1.8 to 0.9); (f) Kyte & Doolittle (Citation1982) (−4.5 kcal/mol to 4.5 kcal/mol); (g) Rose et al. (Citation1985) (0.52 to 0.91); (i) Schauperl et al. (Citation2016) (−15 to −5) and (k) the ΔG values of the surrounding water molecules (cut-off 4 Å) calculated using GIST tool (−4kcal/mol to 1 kcal/mol).
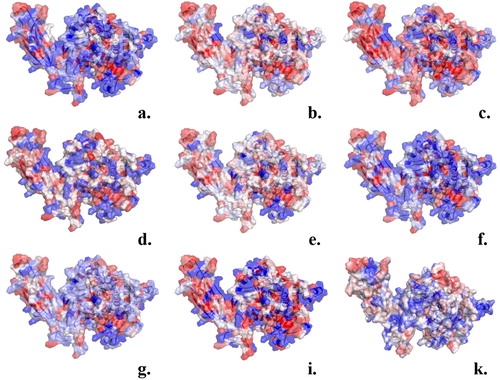
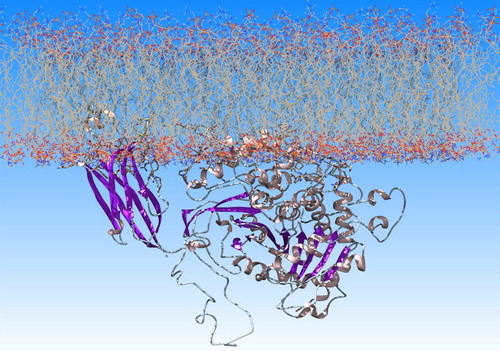
Figure 3. Orientation of GIVA cPLA2 in a lipid bilayer according to DXMS data. The description of the hydrophobic areas (red) of the protein surface was produced by the GIST tool. These areas penetrate the membrane and this is in accordance with the DXMS data.
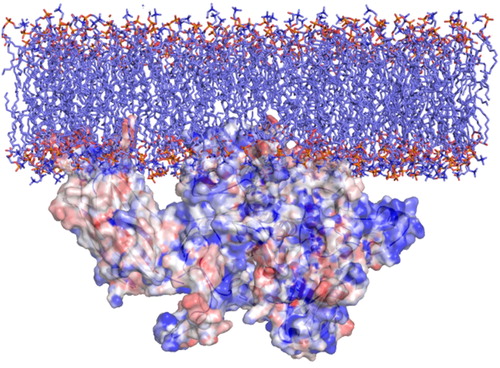
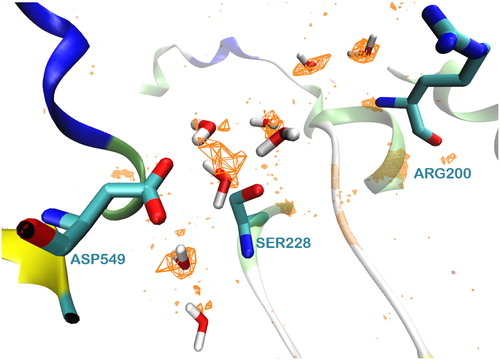
Figure 4. A representative snap shot from the MD simulation of the protein-membrane complex. The entrance of the tunnel (yellow residues) is highlighted with a yellow circle. It leads to the catalytic dyad Ser228/Asp549 shown by the orange arrow. Met468 and Trp464 (yellow) interact with the lipophilic part of lipids (cyan) and Arg467, Lys273 and Arg274 (yellow) with the polar head groups, in accordance with the DXMS data (only some parts of the lipids are represented here (cyan ‘stick’) for clarity reasons).
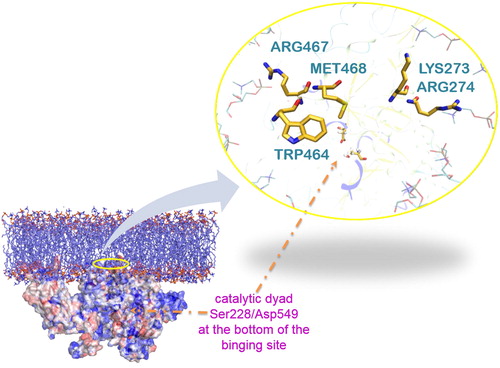
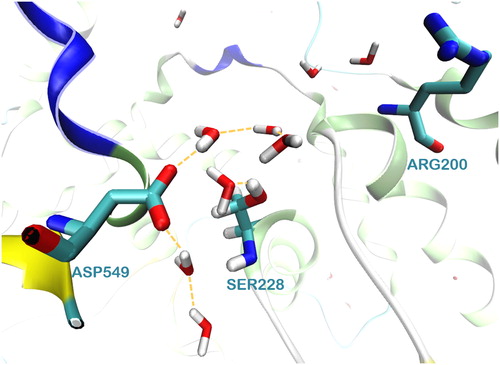
Figure 5. The network of water molecules around the catalytic dyad Ser228/Asp549 of GIVA cPLA2 formed during a 300 ns MD simulation.