Figures & data
Figure 1. Circadian clock. Model of the circadian core oscillator, showing the main core clock genes and transcription/translation feedback loops, as well as clock (controlled) genes (CCGs) that link the molecular clock to oscillating output processes. The core clock proteins are depicted as circles (CLOCK, ARNTL, CRY, PER, NR1D1).
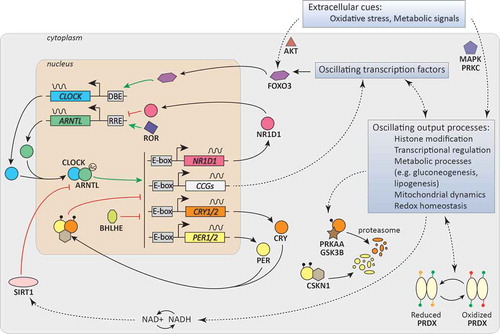
Figure 2. Flowchart of the study population. EOPE: early-onset preeclampsia; LOPE: late-onset preeclampsia; CTRL: uncomplicated controls; FGR: fetal growth restriction; PTB: preterm birth; UCL: umbilical cord leukocytes; HUVEC: human umbilical vein endothelial cells.
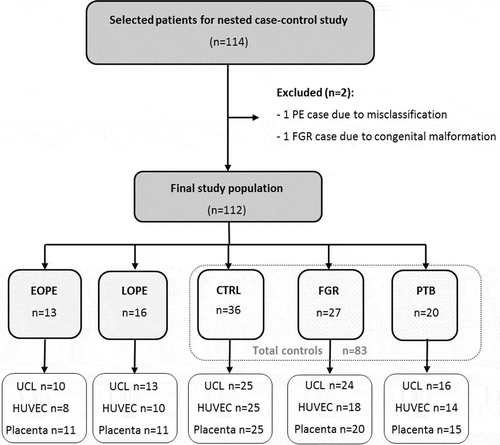
Figure 3. Selection of circadian clock and clock-controlled genes and CpGs. AKT1: gene; V-Akt: murine thymoma viral oncogene homolog1; ARNTL: aryl hydrocarbon receptor nuclear translocator-like; ARNTL2: aryl hydrocarbon receptor nuclear translocator-like 2; BHLHE40: basic helix-loop-helix family, member e40; BHLHE41: basic helix-loop-helix family, member e41; CLOCK: clock circadian regulator; CRTC1: CREB-regulated transcription coactivator 1; CRY1: cryptochrome circadian clock 1; CRY2: cryptochrome circadian clock 2; CSNK1E: casein kinase 1, epsilon; DBP: D site of albumin promoter (albumin D-box) binding protein; FGF21: fibroblast growth factor 21; FOXO1: forkhead box O1; FOXO3: forkhead box O3; GSK3B: glycogen synthase kinase 3 beta; MAPK1: mitogen-activated protein kinase 1; NPAS2: neuronal PAS domain protein 2; NR1D1: nuclear receptor subfamily 1, group D, member 1; NR1D2: nuclear receptor subfamily 1, group D, member 2; PER1: period circadian clock 1; PER2: period circadian clock 2; PER3: period circadian clock 3; PRDX1: peroxiredoxin 1; PRDX2: peroxiredoxin 2; PRDX3: peroxiredoxin 3; PRDX5: peroxiredoxin 5; PRDX6: peroxiredoxin 6; PRKAA1: protein kinase, amp-activated, alpha 1 catalytic subunit; PRKAB1: protein kinase, AMP-activated, beta 1 non-catalytic subunit; PRKACA: protein kinase, CAMP-dependent, catalytic, alpha; PRKCA: protein kinase C, alpha; RORA: RAR-controlled orphan receptor A; RORB: RAR-controlled orphan receptor B; RORC: RAR-controlled orphan receptor C; SIRT1: sirtuin 1; SIRT2: sirtuin 2; STRA13: stimulated by retinoic acid 13; TIMELESS: timeless circadian clock; WEE1, WEE1 G2 checkpoint kinase.
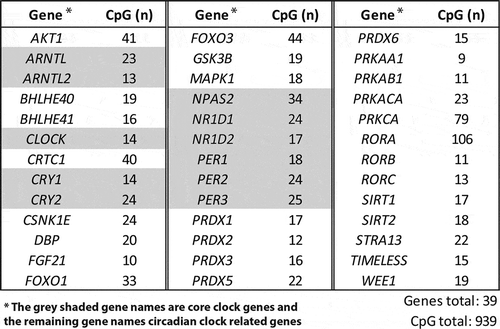
Table 1. Maternal and newborn characteristics.
Figure 4. Differentially methylated CpGs for the three different tissues. An overall ANOVA was performed to test for differentially methylated CpGs (p < 0.05) between all study groups. UCL: umbilical cord leukocytes; HUVEC: human umbilical vein endothelial cells.
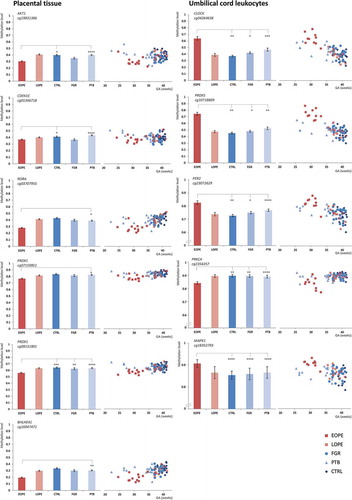
Figure 5. EOPE and LOPE compared to different control groups (ANCOVA). ANCOVA, adjusted for gestational age at delivery, batch effect, and leukocyte count for UCL only was performed. p Values are shown in the columns. EOPE: early-onset preeclampsia; LOPE: late-onset preeclampsia; CTRL: uncomplicated controls; FGR: fetal growth restriction; PTB: preterm birth; UCL: umbilical cord leukocytes; HUVEC: human umbilical vein endothelial cells; n.s.: not significant.
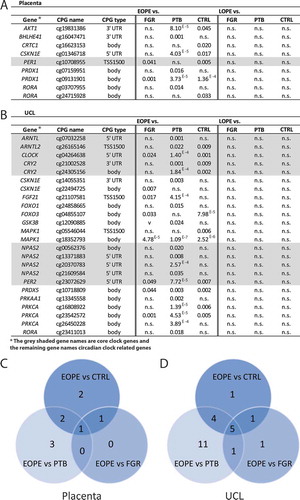
Figure 6. Absolute DNA methylation levels in the EOPE, LOPE and (un)complicated control groups depicted at the gestational age of delivery. EOPE: early-onset preeclampsia; LOPE: late-onset preeclampsia; CTRL: uncomplicated controls; FGR: fetal growth restriction; PTB: preterm birth. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 1E-4, *****p < 1E-5.