Figures & data
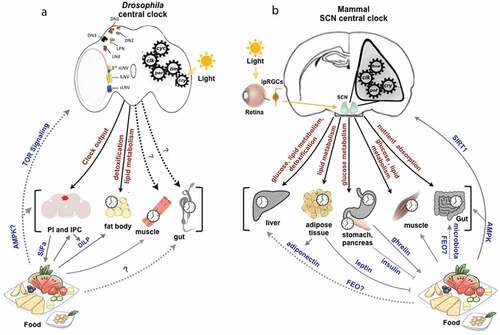
Figure 1. Circadian clock in Drosophila and mammals. (a) Primary and secondary feedback loops in the Drosophila circadian clock. The core clock machinery in Drosophila is composed of an interlocked TTFL with the positive limb CLK/CYC and a negative limb PER/TIM. Circadian photoreceptor CRY mediates the light entrainment of circadian clocks in Drosophila. The secondary feedback loop consists of VRI and PDP1 that inhibit and activate Clk transcription, respectively. (b) Primary and secondary feedback loops in the mammalian circadian clock. In mammals, the TTFL consists of the positive arm CLOCK/BMAL1 and a negative arm PER/CRY. REV-ERB and ROR, inhibit and activate Bmal1 transcription, respectively.
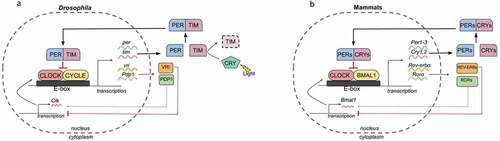
Figure 2. Central and peripheral circadian clocks in Drosophila and mammals.