Figures & data
Figure 1. Overexpression of LINC01232 in ccRCC tissues and cell lines. (A) LINC01232 expression in 535 ccRCC tissues and 72 normal tissues from TCGA database by starBase v3.0 analysis. (B) LINC01232 expression in tumour tissues and normal tissues from 122 ccRCC patients from our study cohort (fold change is 0.89). (C) LINC01232 expression in ccRCC cell lines and normal renal tubular epithelial cell line (fold changes are 2.64, 2.25, 1.98, 2.54 and 1.75, respectively). ***p < .001 vs. ccRCC tissues from TCGA database or normal tissues from 122 ccRCC patients or HK-2. LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
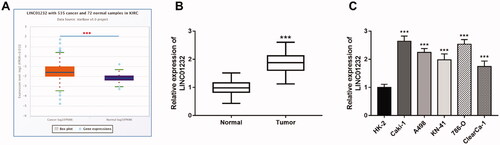
Figure 2. LINC01232 is associated with overall survival of ccRCC patients. (A) According to the analysis by starBase, high LINC01232 was associated with poor overall survival of ccRCC patients (log-rank p = .002). (B) The results of our study cohort revealed that patients with high LINC01232 level had short survival time than the patients with low LINC01232 level (log-rank p = .006). LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
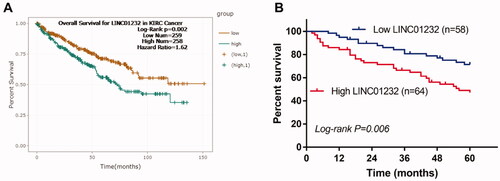
Table 1. Association of LINC01232 with the clinicopathological characteristics of ccRCC.
Table 2. Cox regression analysis results for patients with ccRCC.
Figure 3. Effects of LINC01232 on the proliferation, migration and invasion of ccRCC cells. (A) LINC01232 expression was inhibited by sh-LINC01232 in Caki-1 (fold change is 0.22) and 786-O cells (fold change is 0.25). (B–D) LINC01232 silencing inhibited the proliferation, migration and invasion of Caki-1 and 786-O cells. **p < .01, ***p < .001 vs. Control. sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
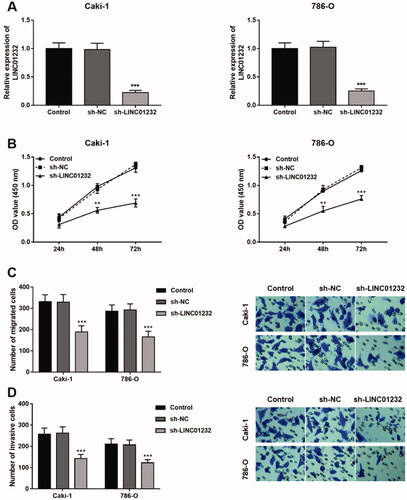
Figure 4. LINC01232 sponges miR-204-5p in ccRCC. (A) The binding site of LINC01232 to miR-204-5p was predicted. (B) LINC01232 was found to be negatively correlated with miR-204-5p in ccRCC by starBase platform (r= −0.220, p < .001). (C) In Caki-1 cells, relative luciferase activity was significantly inhibited by miR-204-5p upregulation in WT-LINC01232 group. (D) LINC01232 expression was downregulated by sh-LINC01232 and was upregulated by pcDNA3.1-LINC01232 (fold changes are 0.21 and 2.91). (E) miR-204-5p levels were promoted by LINC01232 silencing and were suppressed by the overexpression of LINC01232 in Caki-1 cells (fold changes are 1.97 and 0.4). (F) Through starBase platform, miR-204-5p was significantly downregulated in ccRCC. (G) Through the starBase platform, ccRCC patients with low miR-204-5p levels had a poor overall survival (log-rank p < .001). (H) miR-204-5p levels in ccRCC tumour tissues and normal tissues (fold change is 0.39). (I) miR-204-5p levels were negatively correlated with LINC01232 levels in tumour tissues (r= −0.600, p < .001). (J) Patients with low miR-204-5p levels had a poor five-year survival (log-rank p = .002). *p < .05, ***p < .001 vs. Control or ccRCC tissues from TCGA database or normal tissues from 122 ccRCC patients. WT: wide-type; MUT: mutant-type; sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
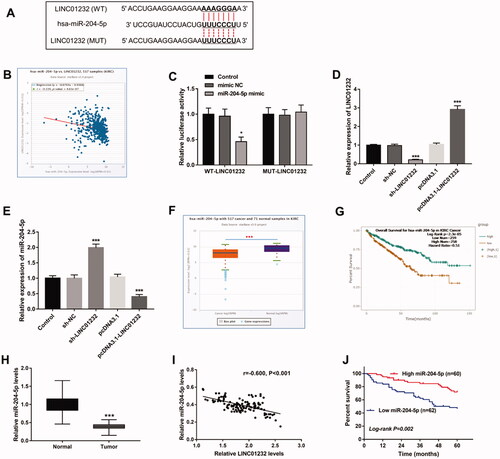
Figure 5. miR-204-5p mediates the effects of LINC01232 on ccRCC cell proliferation, migration and invasion. (A) The upregulation of miR-204-5p caused by LINC01232 silencing was reversed by miR-204-5p downregulation in Caki-1 cells (fold changes are 1.96, 0.31 and 1.10, respectively). (B–D) miR-204-5p downregulation reversed the inhibitory effects of LINC01232 silencing on Caki-1 cell proliferation, migration and invasion. **p < .01, ***p < .001 vs. Control; #p < .05, ##p < .01, ###p < .001 vs. sh-LINC01232. sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
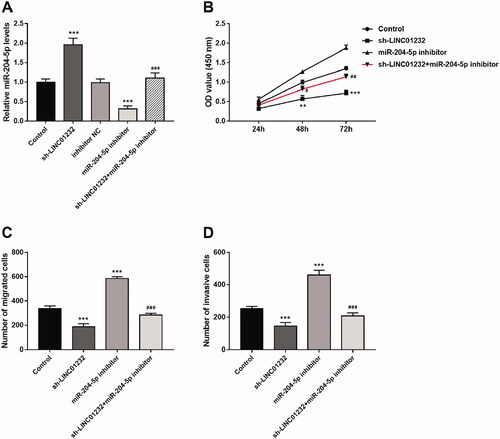
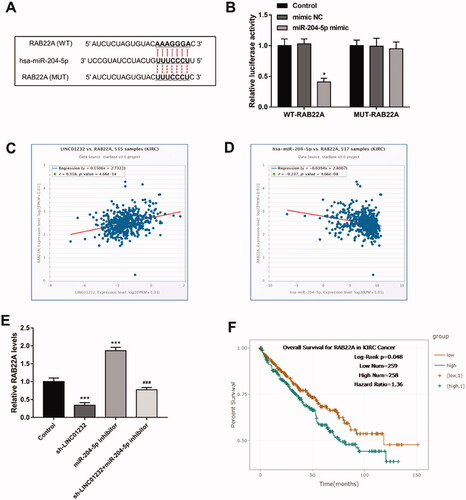
Figure 6. LINC01232 positively regulates RAB22A through sponging miR-204-3p in ccRCC. (A) The binding sites of miR-204-5p and RAB22A were predicted. (B) miR-204-5p upregulation significantly inhibited relative luciferase activity in the WT-RAB22A group. (C–D) Through the starBase platform, in ccRCC, LINC01232 was positively correlated with RAB22A (r = 0.318, p < .001) and miR-204-5p was negatively correlated with RAB22A (r= −0.237, p < .001). (E) miR-204-5p downregulation reversed the suppressive effects of LINC01232 silencing on RAB22A levels in Caki-1 cells (fold changes are 0.34, 1.87 and 0.77, respectively). (F) Through the starBase platform, high RAB22A levels were associated with poor overall survival in ccRCC patients (log-rank p = .048). *p < .05, ***p < .001 vs. Control; ###p < .001 vs. sh-LINC01232. WT: wide-type; MUT: mutant-type; sh: short hairpin; NC: negative control; LINC: long intergenic non-protein coding RNA; ccRCC: clear cell renal cell carcinoma.
Supplemental Material
Download ()Data availability statement
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
