Figures & data
Table 1. Enzyme inhibitory screening of compounds 1 and 2 isolated from E. pulcherrima.
Figure 1. (a) Chemical structures of isolated compounds from E. pulcherrima. (b) Illustration of predicted docked poses of compounds 1 and 2 (indicated by green colour sticks) in the binding pocket of urease (a), tyrosinase (b), bovine serum albumin (c), and phosphodiesterase-I (d). All predicted conformations were created at the binding site of crystal structures, whereas existing co-crystallized compounds are bonded in the active site.
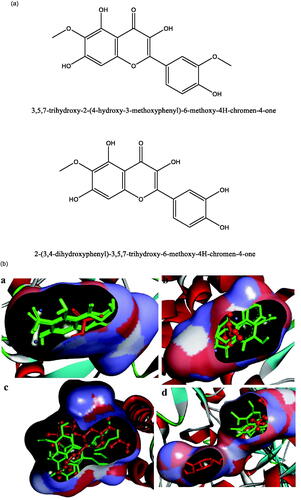
Figure 2. Interactions of compound 1 with the binding pocket of urease enzyme in 2D (a) and 3D (b). Hydrophobic interactions are depicted as half-moons, whereas dotted green lines with distances in angstrom represent hydrogen bonding.
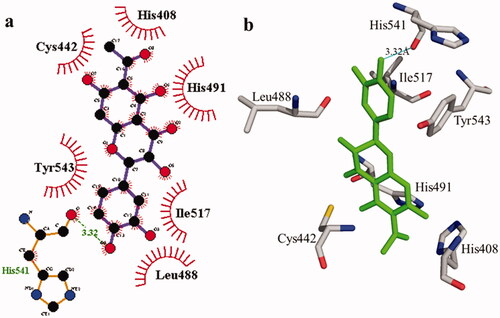
Table 2. Molecular docking details of isolated compounds 1 and 2 and against urease and tyrosinase enzymes.
Table 3. Docking details of BSA and phosphodiesterase-I receptors with compounds 1 and 2.
Figure 3. The interactions of the urease enzyme with compound 2 are determined by binding residues. Compound 2 interacts with the active site in 2D (a) and 3D (b).
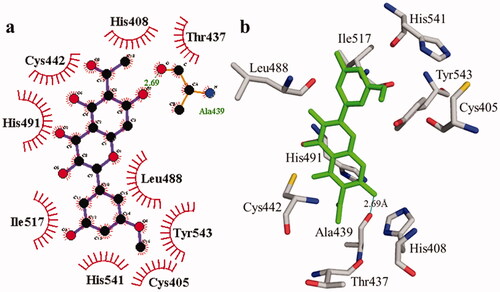
Figure 4. The interactions of the tyrosinase enzyme with compound 1 are determined by binding residues. Detailed interactions of compound 1 with active sites of the enzyme in 2D (a) and 3D (b).
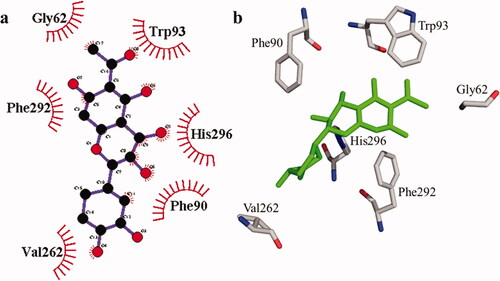
Figure 5. The interactions of the tyrosinase enzyme with compound 2 are determined by binding residues. Detailed interactions of compound 2 with active sites of the enzyme in 2D (a) and 3D (b).
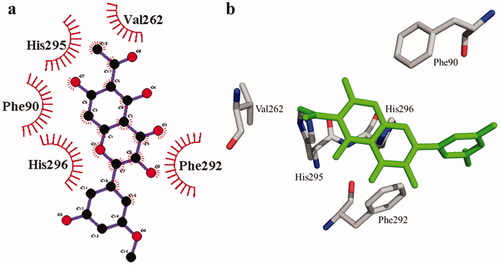
Figure 6. The interactions of bovine serum albumin with compound 1. Detailed interactions of compound 1 with active sites of the enzyme in 2D (a) and 3D (b).
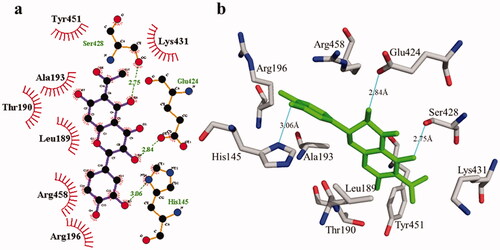
Figure 7. Detail interactions display of compound 2 with bovine serum albumin. Detailed interaction of compound 2 with the active site in 2D (a) and 3D (b).
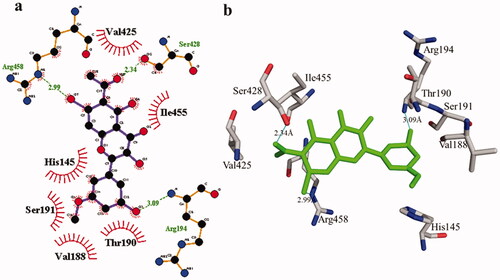
Figure 8. Phosphodiesterase-I interactions with compound 1 are mediated by binding residues. Detailed interaction of compound 1 with the active site is shown in 2D (a) and 3D (b).
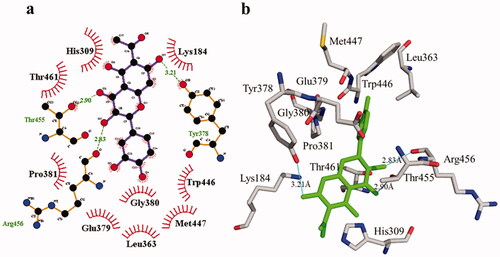
Figure 9. Phosphodiesterase-I interactions with compound 2 are mediated by binding residues. Detailed interaction of compound 2 with the active site in 2D (a) and 3D (b).
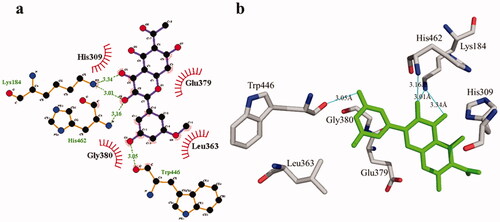
Table 4. Prediction of ADMET properties of compounds 1 and 2.
Data availability statement
The data based on the results of the current study are obtained from the corresponding authors upon request.
