Figures & data
Table 1. Characteristics of the sample cohorts whose serum was used in this study.
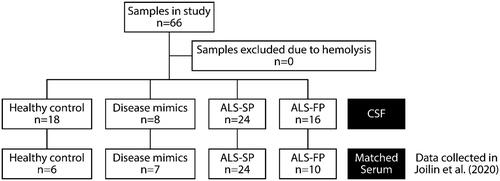
Figure 2. RNA-seq shows ncRNA may be dysregulated in CSF from ALS patients. (A) Workflow of the experiment from sample to sequencing data. (B) Summary of the number of aligned reads that were annotated and the ncRNA species they aligned to. (C) MA plots of ncRNA dysregulation in slow- (ALS-SP) and fast-progression (ALS-FP) ALS patients compared to healthy controls, and ALS-FP compared to ALS-SP. Dots in red were significantly dysregulated compared to healthy control (horizontal dotted line representing p < 0.05).
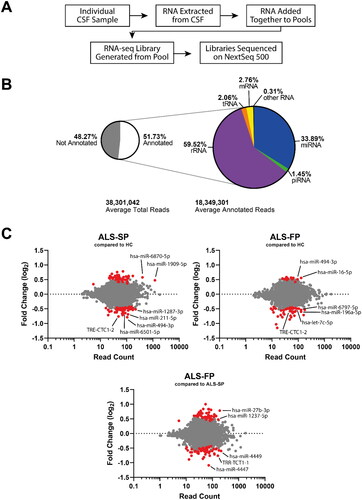
Figure 3. RT-qPCR profiling of ncRNA biomarker candidates in CSF from RNA-seq data. Those ncRNA transcripts that showed normal amplification during RT-qPCR across a majority of samples showed no significant dysregulation compared to the average of the healthy control samples. ALS-FP: fast-progressing ALS, ALS-SP: slow-progressing ALS; relative expression ± SD; normalised to geometric average of hsa-miR-9-5p and hsa-miR-501-3p; one-way ANOVA with Tukey’s.
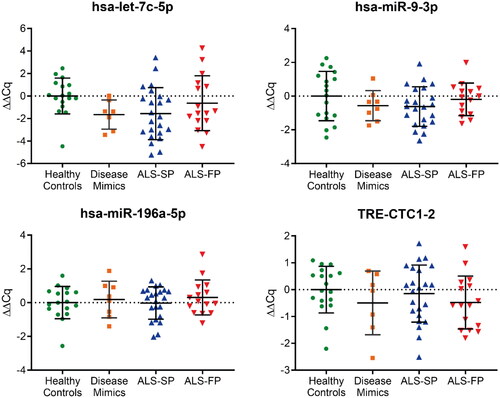
Figure 4. Expression of serum-based ncRNA biomarker candidates in CSF in the BioMOx cohort. (A) No significant dysregulation of serum-based ncRNA biomarker candidates from Joilin et al. [Citation7] were detected in CSF compared to the healthy controls. (B) No significant correlation in expression between CSF and serum in matched patient samples in the BioMOx cohort across all groups. Data from serum samples were collected as part of the study in Joilin et al. [Citation7]. ALS-FP: fast-progressing ALS, ALS-SP: slow-progressing ALS; relative expression ± SD; normalised to geometric average of hsa-miR-9-5p and hsa-miR-501-3p; relative expression: one-way ANOVA with Tukey’s; correlation: Pearson’s correlation.
![Figure 4. Expression of serum-based ncRNA biomarker candidates in CSF in the BioMOx cohort. (A) No significant dysregulation of serum-based ncRNA biomarker candidates from Joilin et al. [Citation7] were detected in CSF compared to the healthy controls. (B) No significant correlation in expression between CSF and serum in matched patient samples in the BioMOx cohort across all groups. Data from serum samples were collected as part of the study in Joilin et al. [Citation7]. ALS-FP: fast-progressing ALS, ALS-SP: slow-progressing ALS; relative expression ± SD; normalised to geometric average of hsa-miR-9-5p and hsa-miR-501-3p; relative expression: one-way ANOVA with Tukey’s; correlation: Pearson’s correlation.](/cms/asset/f8451e3a-735a-471c-b2c5-71219ae795e6/iann_a_2138530_f0004_c.jpg)
Supplemental Material
Download MS Excel (648.3 KB)Supplemental Material
Download MS Word (224.4 KB)Data availability statement
The data that support the findings of this study are available from the corresponding author, MH, upon reasonable request.