Figures & data
Figure 1. Resistance mechanisms to Top I inhibitors in cancer. (a) Overexpression of efflux transporters (PgP, ABCB1, MRP, ABCC and BCRP) can reduce the cellular abundance of Top I inhibitors. (b–e) Mutations in Top I gene, reduced Top I expression and phosphorylation levels and reduced sumoylation of Top I enzyme itself can decrease the affinity of Top I inhibitors to their target. (f) Overexpression of tyrosyl-DNA phosphodiesterase 1 (TDP-1) can increase the rate of cleavage of the covalent linkage between stabilized Top1 with DNA and reverses the formation of cleavage complex. All these events lead to the development of resistance to Top I inhibitors. PgP: P-glycoprotein; ABCB1: ATP Binding Cassette Subfamily B Member 1; MRP/ABCC: multidrug resistance related protein; BCRP/ABCG2: breast cancer resistance protein; Top I: topoisomerase I; TDP-1: tyrosyl-DNA phosphodiesterase 1.
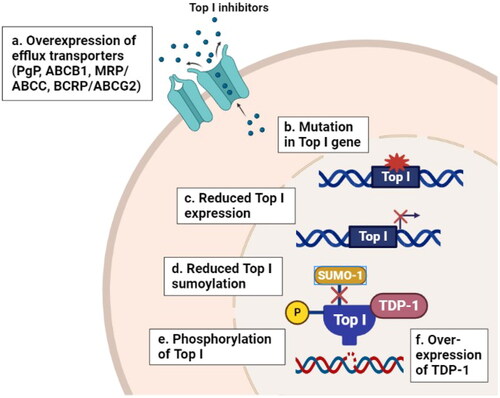
Figure 2. Effect of DNA hypomethylation on the response of cancer cells to CPTs. (A) mechanisms by which hypomethylation can increase sensitivity to CPTs including: decreased expression of Bcl-2 anti-apoptotic protein, increased number of replication forks in early S phase, demethylation of the Top I promoter or indirect stimulation of Top I expression and restored expression of DEXI protein. (B) Mechanisms by which hypomethylation can decrease sensitivity to CPTs including protection of newly synthesized DNA from cleavage complex stabilization and DNA fragmentation, elevated expression of UGT1A1 metabolizing enzyme which is responsible for irinotecan’s inactivation and elevated expression of WRN helicase enzyme.
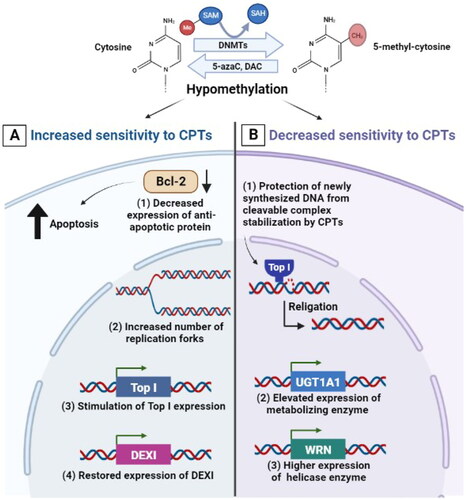
Figure 3. Effect of targeting histone modifying enzymes on cellular response to CPTs. Gene deletion or inhibition of HATs, inhibition of histone deacetylation by HDACi or inhibiting BET, KDMs and EZH2 result in higher DNA damage levels, ROS generation, increase in caspase 3/7 activity and apoptosis induction. KDMi: histone lysine demethylase inhibitor; EZH2i: histone-lysine N-methyltransferase enzyme enhancer of zeste homolog 2 inhibitor; CPTs: camptothecins; Top I: topoisomerase I; HATi: histone acetyltransferase inhibitor; HDACi: histone deacetylase inhibitor; BETi: bromodomain and extraterminal domain inhibitor; DSBs: double strand breaks; ROS: reactive oxygen species.
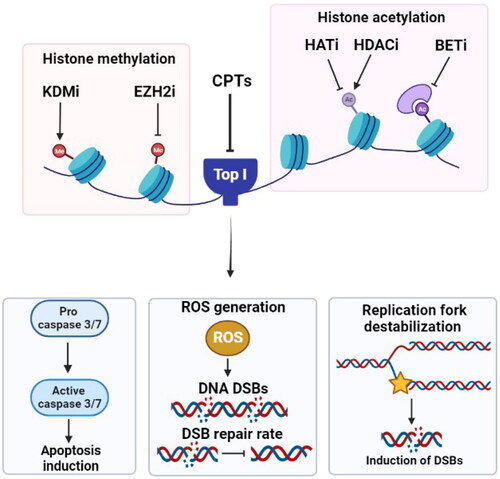
Figure 4. The involvement of miRNAs and lncRNAs in the response to Top I inhibitors. (A) miR-23a and miR-139 inhibit Top I expression. Thus, curbing their expression will in turn augment the enzyme’s expression. (B) Higher levels of miR-519c can lead to decreased ABCG2 expression leading to higher sensitivity to CPTs. (C) lncRNAs participate in maintaining genomic stability, cell survival and higher resistance to CPTs. Absence of lncRNAs can lead to persisitence of Top I cleavage complexes, increased chromosomal defects and better response to CPTs.
Table 1. Clinical trials for combination of epigenetic drugs and Top I inhibitors.
Data availability statement
Data sharing not applicable – no new data generated.
