Figures & data
Table I. The biochemistry of apoB-containing lipoproteins: A: Abbreviations; and B: Review referrals—topics.
Figure 1. Secretion of apoB-containing lipoproteins. The schematic drawing illustrates the cellular components involved in the secretion of apoB-containing lipoproteins into the serum. The apoB polypeptide chain either apoB-100 or apoB-48 is synthesized by ribosomes attached to the rough ER and cotranslationally moved into this secretion system. The NH2-terminal segment of undefined length forms a complex with the heterodimeric MTP:PDI complex. MTP and nascent apoB begin loading with lipid. As the nascent lipoproteins increase their lipid content and apoB continues its folding process, more mature form(s) of appear and are transferred to the Golgi where secretion vesicles are formed and released from the cell. LMDs are defined as some yet undefined lipid microdomains produced by MTP or the MTP:PDI complex. A single LMD is metastable and would contain only a small portion of the overall lipid load. Formed by fragmentation of lipid droplets or membrane segments, LMDs may contain phospholipids, triacylglycerols, and various forms of cholesterol. Their unique property would be a covering having both hydrophobic and polar surfaces. Although not explicitly shown in the schematic drawing, a sorting system removes misfolded proteins from the secretory process. This hypothetical assembly route is similar to that proposed by Olofsson et al. Citation62.
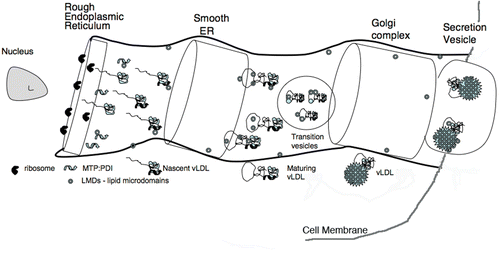
Figure 2. The dimer of lipovitellin. The cartoon model depicts the crystal structure of lamprey lipovitellin. It is thought to be a homolog of the NH2-terminal of apoB and the intact MTP proteins. The LV dimer has 2-fold rotational symmetry, with the position of the 2-fold axis indicated by the elliptical black mark in the center of the diagram. Each subunit, labeled 1 and 2, is composed of multiple domains illustrated by the colors red, blue, and green (or shades of gray) and the letters a, b, c. The green domains labeled 1a and 2a include residues from the NH2-terminal region. The domains shown in red (1b and 2b) are comprised of a double layer of α-helices. The blue domains, labeled 1c and 2c, consist of mainly of β-strands forming several sheets that surround the lipid-binding cavity.
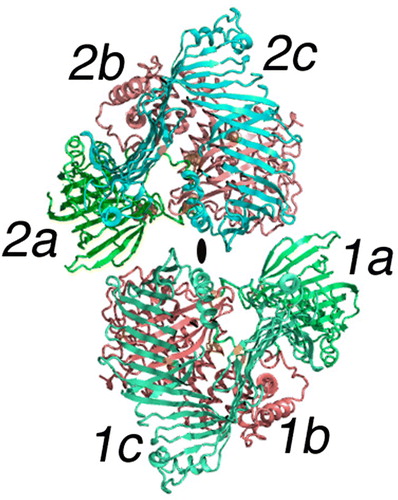
Figure 3. Lipid molecules attached to a subunit of LV. The cartoon is color-coded the same as but shows only a single subunit of LV in an orientation where the major lipid-containing region is visible. The green portion labeled 1a is again the NH2-terminal domain; 1b in red is the helical domain; 1c is the β-sheet domain containing the lipid core. Two unique lipid-binding sites are labeled L1 and L2 and are partially hidden but visible as black spherical nets. Other lipid molecules or parts thereof observed in the crystallographic studies are evident in the lipid cavity as black bonds. L2 is a phospholipid as reported in the original study Citation14. The center of this segment of the LV molecule is labeled lipid core. Although not containing any visible electron density in the crystal structure, other experiments establish this location as that containing most of the bound lipid; for LV, 15% w/w is lipid, mostly phospholipid but neutral lipid as well.
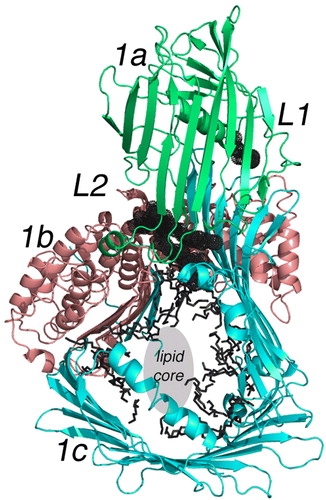
Figure 4. The binding site of phospholipid L2 in lipovitellin. The drawing is a cartoon representing the X-ray crystal structure of LV in a region near the bound phospholipid L2. L2 is visible as a black stick model surrounded by dot spheres. The binding site occurs between β-strands of the a- and c-domains. Note how the two methylene chains of the phospholipid are positioned directly between the two β-sheets, one from the a-domain and another from the c-domain. It is hypothesized that movement at this junction could change the volume of the lipid core, and hence the binding of L2 is a signal for further lipid loading to occur. Removal of L2 could cause contraction of the main lipid cavity and expulsion of some of the accompanying lipid.
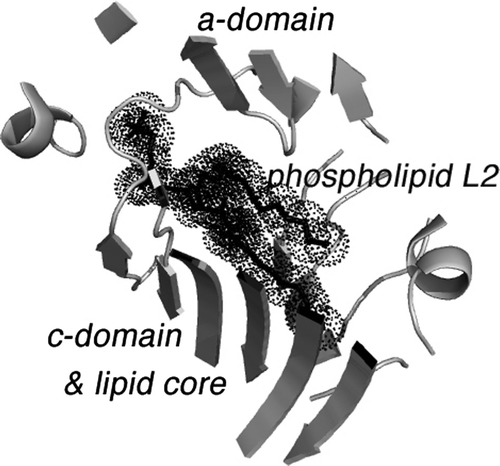
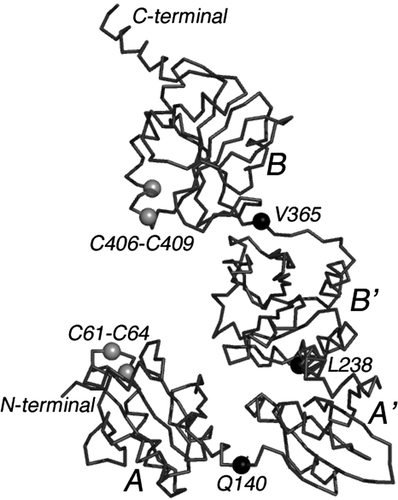
Figure 5. The backbone structure of yeast PDI. The Cα model of yeast protein disulfide isomerase is shown (PDB accession—2b5e) Citation64. Both the NH2- and COOH-terminals are labeled. As noted in the text, PDI consists of approximately 130 residue domains labeled A, B, B' and A' in the accompanying drawing. Like beads on a string, the four domains are interconnected by what appears to be relatively flexible loops marked approximately in the middle by black balls (Q140, L238, and V365). Disulfide redox centers are present only in domains A and A'. They are labeled as C61-C64 and C406-C409.