Figures & data
Figure 1. Overview of the lipoprotein isolation and measurement procedures for Data set I. Plasma C and plasma TG are measured, and identical samples are also handled through two different procedures. In procedure A the HDL-C is measured after VLDL ultracentrifugation and precipitation of the apoB-containing lipoprotein particles. In procedure B, sequential ultracentrifugation is applied to physically isolate the individual VLDL, IDL, LDL, and HDL fractions. ▵HDL-C(AB) denotes the difference of the measured HDL-C in procedures A and B. (Key for the abbreviations: C = cholesterol; TG = triglycerides; VLDL = very-low-density lipoprotein; IDL = intermediate-density lipoprotein; LDL = low-density lipoprotein; HDL = high-density lipoprotein; UCF = ultracentrifugation.) See also Supplementary .
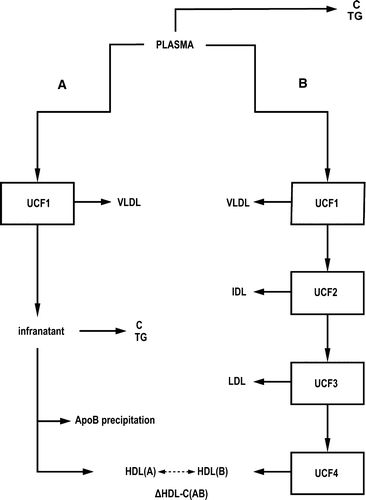
Table I. Characteristics of the lipid and apolipoprotein measures in Data set I.
Figure 2. Correlation plots for the cross-validation results for the ANN-estimated variables (Data set I; ). The dashed lines represent the actual regression lines, and the solid lines identify the 1:1 line. An r stands for the Pearson correlation coefficient between the measured and estimated variables. The equations for the regression lines are: A) y = 0.93x + 0.05; B) y = 0.61x + 0.11; C) y = 0.89x + 0.36; D) y = 0.88x + 0.14; E) y = 0.90x + 0.17; and F) y = 0.92x + 0.10.
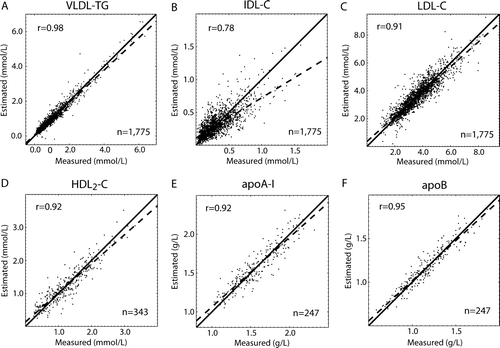
Figure 3. Bland-Altman plots for the cross-validation results for the ANN-estimated variables (Data set I; ). The solid line in the middle represents the mean bias and the two others the mean±2 SD. The dashed lines represent the regression lines for the biases. A) Bias as a function of VLDL-TG: y = − 0.012x + 0.015 with r=0.054; B) bias as a function of IDL-C: y = − 0.17x + 0.055 with r=0.23; C) bias as a function of LDL-C: y = − 0.019x + 0.063 with r=0.042; D) bias as a function of HDL2-C: y = − 0.031x + 0.036 with r=0.079; E) bias as a function of apoA-I: y = − 0.027x + 0.050 with r=0.068; and F) bias as a function of apoB: y = − 0.022x + 0.034 with r=0.069.
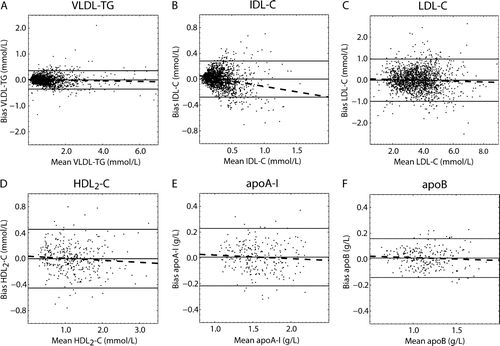
Figure 4. Correlation and Bland-Altman plots for the (IDL + LDL)-C estimates calculated via the Friedewald formula (F-LDL-C) and via the (cross-validated) ANN regression models. The dashed lines in the correlation figures (A and B) represent the regression lines, and the solid lines denote the 1:1 lines. An r stands for the Pearson correlation coefficient between the measured and estimated variables. In the Bland-Altman plots (C and D) the solid line in the middle represents the mean bias and the two others the mean±2 SD. The dashed lines represent the regression lines for the biases. The equations for the regression lines are: A) y = 0.90x + 0.40; and B) y = 0.94x − 0.22. C) Bias as a function of (IDL + LDL)-C: y = − 0.04x + 0.18 with r=0.11; and D) bias as a function of F-LDL-C: y = 0.03x − 0.53 with r=0.070.
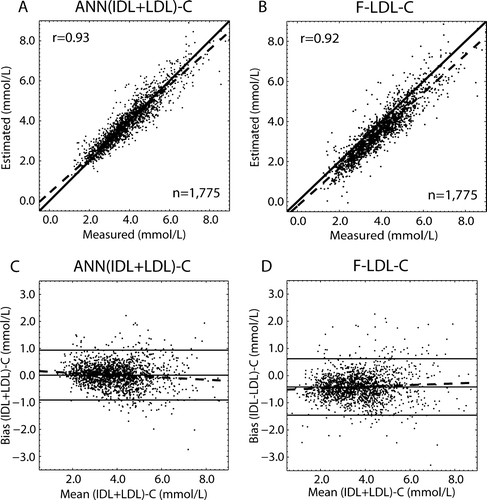
Table II. The distributions of 14 plasma lipid and lipoprotein measures in Data set II consisting of 4,084 patients with type 1 diabetes Citation[20], Citation[25]. The presented Cox hazard regression coefficients and hazard ratios for all-cause mortality are adjusted for age and sex.