Figures & data
FIGURE 1 A fragment of the annotation path for CDC28 in GO molecular function. Indentation on a line denotes refinement of a parent term on the previous line by the child term; in this example, every child has an “is_a” edge to its parent. The numbers in brackets indicate the number of gene products annotated to the GO term. Note the multiple inheritance from the parents on lines 5 and 9 to the term GO:0004672 on lines 6 and 10 and its descendants. CDC28 is annotated to the term GO:0004693 on lines 6 and 12.

TABLE 1 Tests on GO Categories Where One is More General Than the Other are Not Independent—See Text for Details
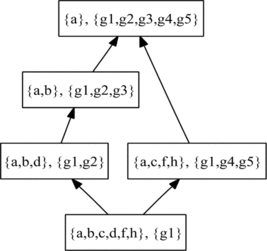
FIGURE 2 Example of a DAG-structured ontology (such as GO, but with a single edge type) and associated objects (such as genes) annotated to its terms.
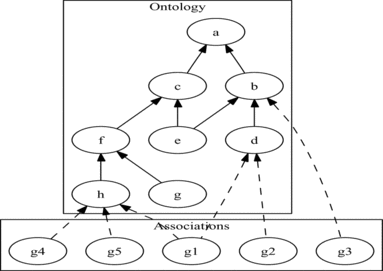
TABLE 2 Coverage Matrix for the Example in Figure
FIGURE 5 Web page to combine gene ontology data with other datasets. Along top of the page are tabs to select a dataset type, such as microarray data or interactions. On the left-hand side, the user can select the mode of activity, such as uploading new data, browsing a dataset, or constructing an integrated set of multirelational data. On the right-hand side are shown the current components of the dataset being constructed. In this example, the GO biological process ontology is being integrated with oxidative stress data.
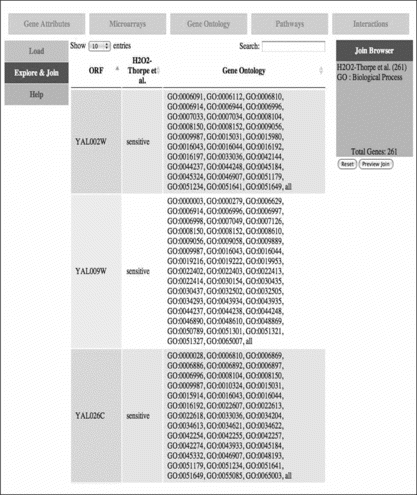
FIGURE 6 Accuracy of predicting protein expression given six microarray datasets (see text for details). The dotted horizontal line shows the baseline accuracy of 61% obtained by simply predicting the majority class for all genes.
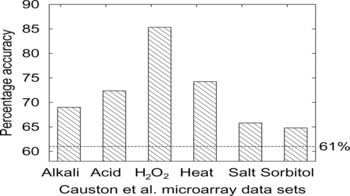
FIGURE 7 Accuracy of predicting protein expression given cellular component (C), molecular function (F), and biological process (P) subontologies of the gene ontology. Four feature selections and construction methods are compared for each subontology (see text for details). The dotted horizontal line shows the baseline accuracy of 61% obtained by simply predicting the majority class for all genes.
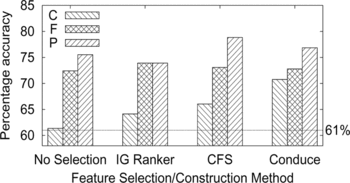
FIGURE 8 A decision tree for protein induction repression learned with gene ontology features. Ovals are attribute tests (“Peroxide t” means microarray data at time t), classifications are at leaves. See text for details.
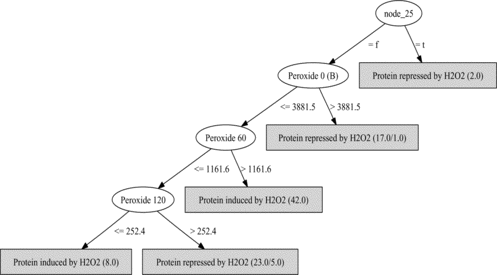
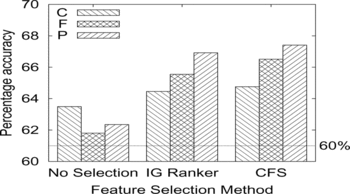
FIGURE 9 Accuracy of predicting general vs. specific deletant sensitivity to multiple stresses given cellular component (C), molecular function (F), and biological process (P) subontologies of the gene ontology. Three feature selection and construction methods are compared for each subontology (see text for details). The dotted horizontal line shows the baseline accuracy of 60% obtained by simply predicting the majority class for all genes.