Figures & data
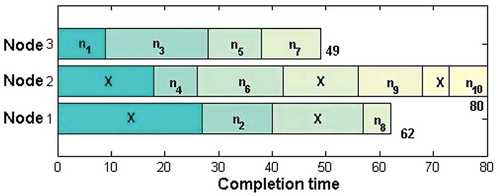
Figure 1. (a) Solution representation. (b) Solution for the problem of 13 tasks and 3 computing nodes. (c) Mapping of tasks with computing nodes for the solution given in (b).

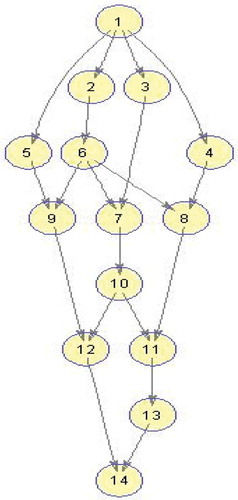
Figure 2. An example DAG(Topcuoglu, Hariri, and Wu Citation2002).
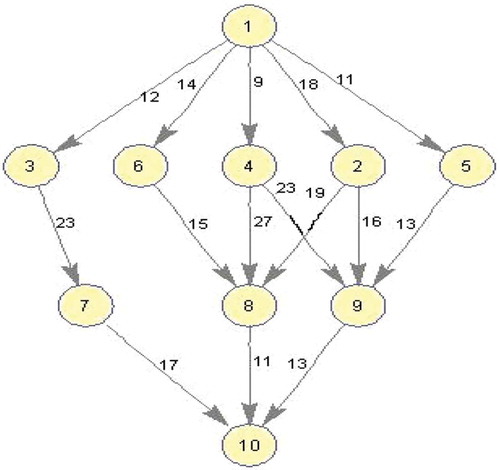
Table 1. Computation cost matrix for random DAG.
Table 2. Upward rank of the tasks.
Table 3. Execution order of tasks.
Figure 3. Explanation of different neighborhood structures: (a) Initial solution, (b) Weightedmakespan–InsertionMove, (c)Makespan-InsertionMove, (d) BestInsertionMove, (e) SwapMove, (f) InsertionMove, (g) InsertionMove, (h) InsertionMove, and (i) InsertionMove.
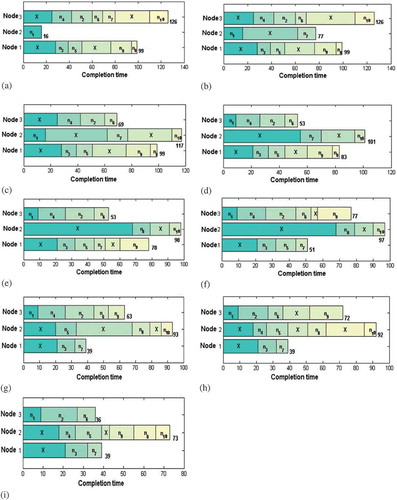
Figure 5. Performance results on random graphs. (a) Average Makespan with respect to DAG size. (b) Average Makespan with respect to CCR.
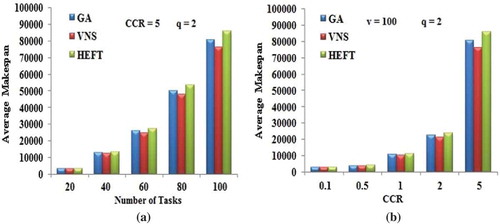
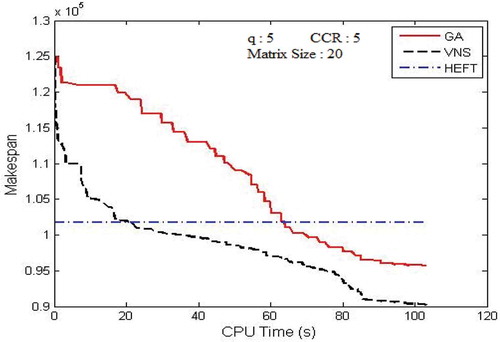
Figure 7. Performance results on Gaussian elimination DAG. (a) Average Makespan with respect to CCR. (b) Average Makespan with respect to number of computing nodes.
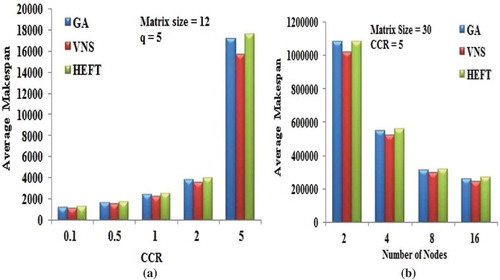
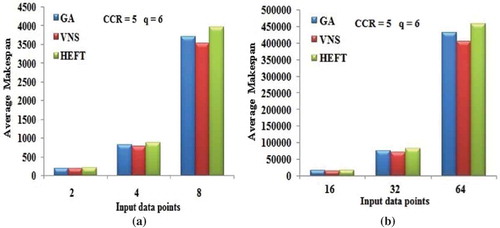
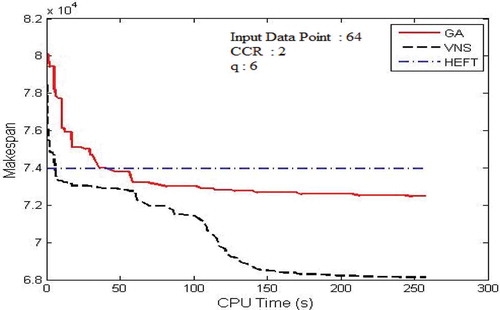
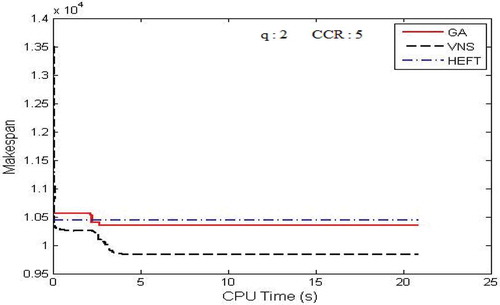
Figure 11. Performance results on FFT DAG. (a) Average Makespan with respect to CCR. (b) Average Makespan with respect to number of computing nodes.
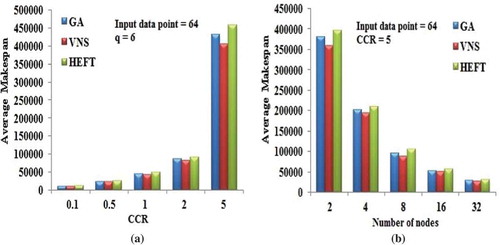
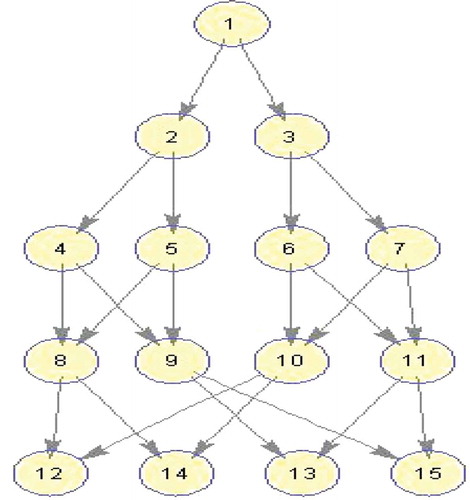
Figure 12. Molecular Dynamics DAG (Topcuoglu, Hariri, and Wu Citation2002).
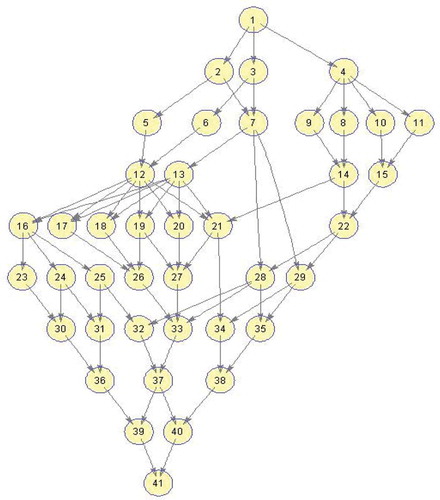
Figure 13. Performance results on Molecular Dynamics DAG. (a) Average Makespan with respect to CCR. (b) Average Makespan with respect to number of computing nodes.
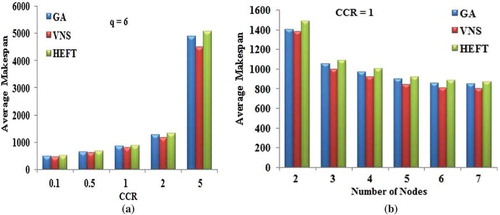
Table 4. Percentage improvement for random graphs with number of tasks.
Table 5. Percentage improvement for random graphs with different CCRs.
Table 6. Percentage improvement for Gaussian elimination DAG with different CCRs.
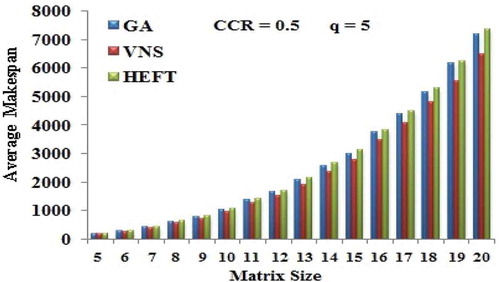
Table 7. Percentage improvement for various matrix sizes of Gaussian elimination DAG.
Table 8. Percentage improvement for Gaussian elimination DAG with the number of nodes.
Table 9. Percentage improvement for FFT DAG with different CCRs.
Table 10. Percentage improvement for FFT DAG with number of nodes.
Table 11. Percentage improvement for various input data points of FFT DAG.
Table 12. Percentage improvement for molecular dynamics DAG with the value of CCR.
Table 13. Percentage improvement for molecular dynamics DAG with number of nodes.