Figures & data
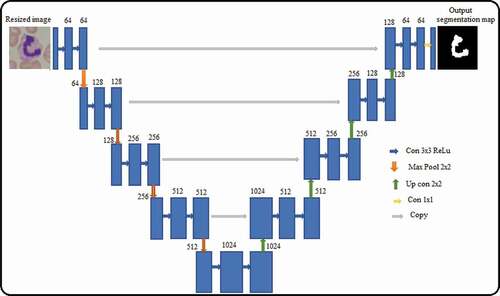
Figure 1. Proposed system architecture for leukemia detection.
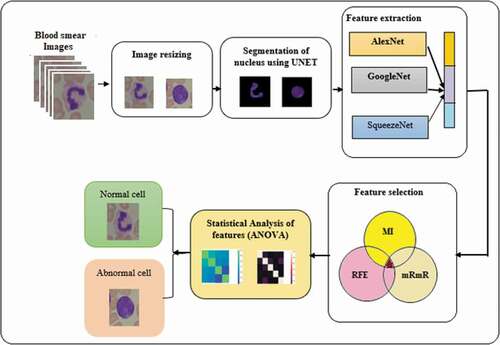
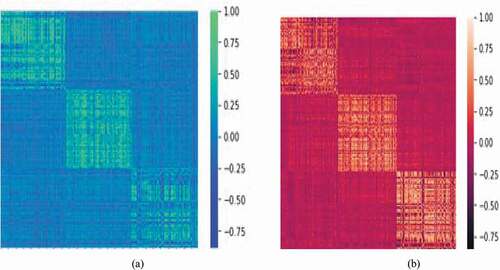
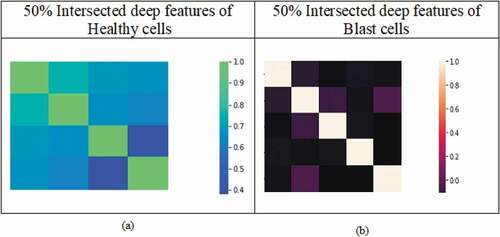
Figure 7. Heat map representation of selected features for healthy cells and blast cells: (a) MI (mutual information), (b) mRmR (minimum recursive maximal relevance) and (c) RFE (recursive feature elimination) based methods.
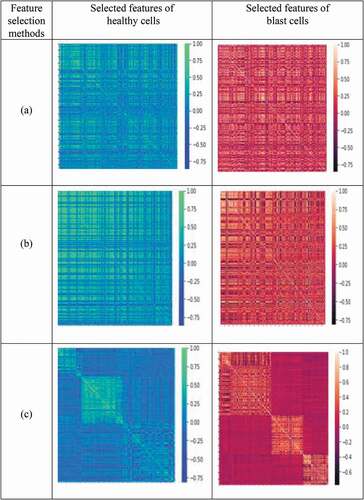
Table 1. Significance of features using ANOVA test
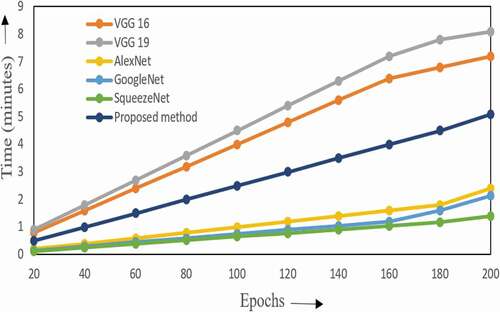
Table 2. Performance analysis of the proposed system with other recent deep learning networks
Table 3. Performance comparison of the proposed method with other existing methods using the ALL–IDB2 dataset