Figures & data
Figure 1 Creatinine clearance of rats exposed to the novel cold I/R model (40 min cold preservation + 18 hr reperfusion). Values are expressed as mean ± SEM (n = 5). *p < 0.05 compared with sham-operated (nephrectomy alone) rats.
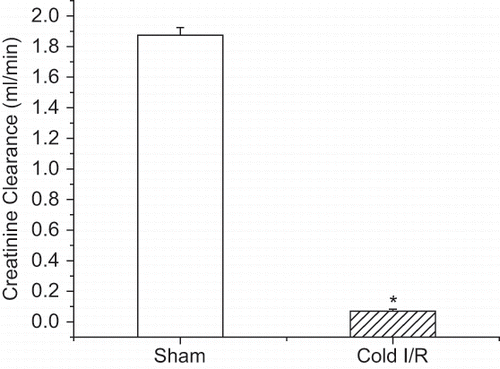
Figure 2 Representative micrographs of PAS (top) and nitrotyrosine (1:500)-stained (bottom) renal sections from rats exposed to cold I/R (40 min cold preservation + 18 hr reperfusion). Experiments were repeated three times with similar results (400 × magnification).
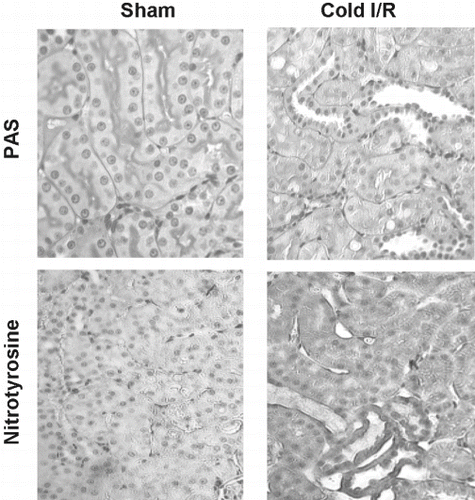
Figure 3 A: MnSOD activity of renal homogenates using the cytochrome c reduction method from rats exposed to cold I/R (40 min cold preservation + 18 hr reperfusion). Values are expressed as mean ± SEM, n = 5. *p < 0.05 compared with sham-operated rats. B: MnSOD Western blot analysis of identical renal homogenates used in panel A. Renal proteins (25 μg) were separated on a 12% SDS-PAGE and blotted with a MnSOD polyclonal antibody (Upstate Biotechnology 1:1000). Blot is representative of three separate experiments.
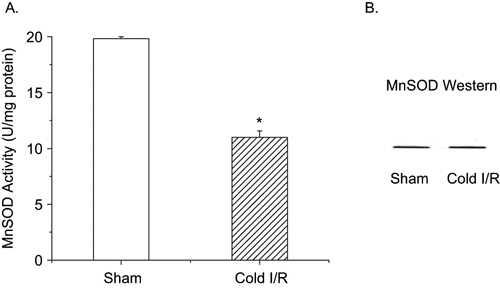
Figure 4 ATP levels of renal extracts following renal cold I/R. Values are expressed as percent change from sham animals (set as 100%) ± SEM, n = 5. *p < 0.05 compared with sham rats.
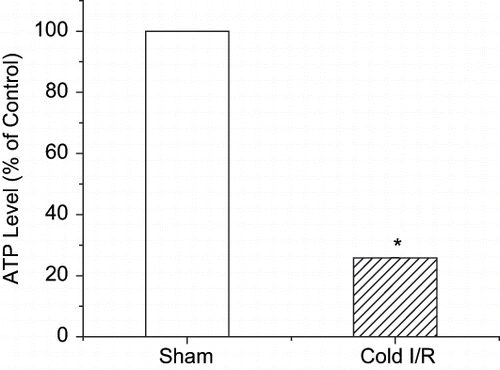
Figure 5 Mitochondrial complex activity assays. Renal mitochondria were prepared from control rats (white bars) or from rats exposed to cold I/R (40 min cold preservation + 18 hr reperfusion-hatched bars). Values are expressed as mean ± SEM, n = 5. *p < 0.05 compared with sham-operated rats.
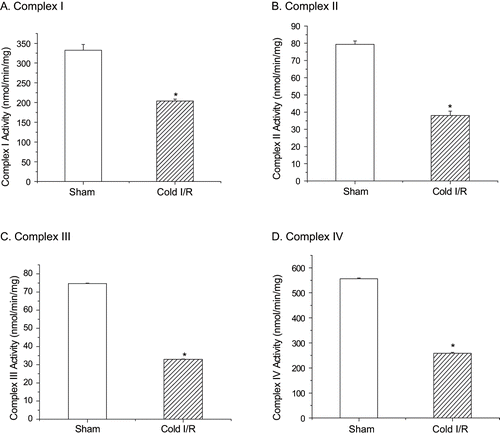
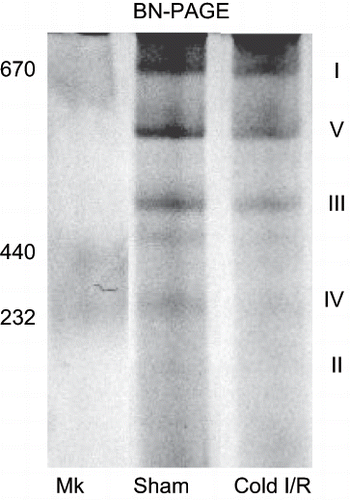
Figure 6 Blue native gel electrophoresis (BN-PAGE) showing alterations in mitochondrial complex proteins following cold I/R (40 min cold preservation + 18 hr reperfusion). Renal mitochondria (80 μg) isolated from rat kidney tissue were electrophoresed (6% native), stained, and destained. The roman numerals represent the migration of mitochondrial electron transport complexes (I-NADH Dehydrogenase; V-ATP synthase, III-Ubiquinol-Cytochrome c Oxidoreductase). The gel is representative of three separate experiments. Mk = molecular weight markers.