Figures & data
Table 1. Primer sequences.
Figure 1. Circ-ITCH overexpression alleviates kidney injury, oxidative stress, and inflammation in lipopolysaccharide (LPS)-induced sepsis mice. (A) Levels of serum creatinine (SCr) and blood urea nitrogen (BUN) were detected by corresponding kits. (B) Levels of neutrophil gelatinase-associated lipocalin (NGAL) and kidney injury molecule-1 (Kim-1) in the urine of mice were detected by ELISA kits. (C,D) Hematoxylin and eosin (H&E) and periodic acid-Schiff staining were used to determine kidney injury (scale bar = 20 µm). (E) H2DCFDA probe was used to detect the reactive oxygen species (ROS) level in kidney tissues of LPS-induced sepsis mice (scale bar = 20 µm). (F) Levels of malondialdehyde (MDA), and superoxide dismutase (SOD) in kidney tissues and interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α in serum were detected by corresponding ELISA kits. LPS-induced sepsis mice were transfected with oe-negative control (NC) or oe-ITCH (ITCH overexpression). *p < 0.05, **p < 0.01.
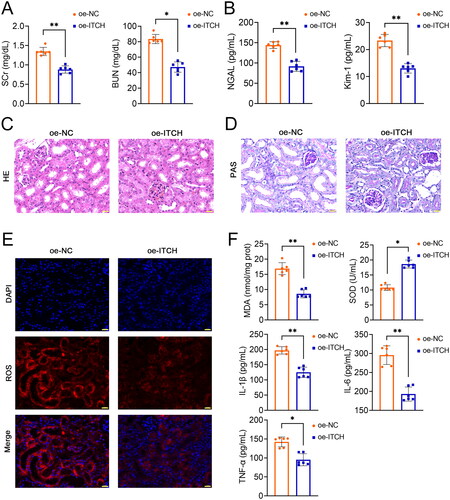
Figure 2. Circ-ITCH protects mitochondrial function of kidney tissues in lipopolysaccharide (LPS)-induced sepsis mice. (A) Adenosine triphosphate (ATP) levels and mitochondrial complexes I/II/III/IV activities in kidney tissues was determined using the corresponding assay kits. (B) Transmission electron microscopy was used to identify mitochondrial morphology in kidney tissues (scale bar = 1 µm). (C-D) RT-qPCR was used to determine the mtDNA copy number and the relative expression of DRP-1 and OPA-1 in kidney tissues. (E) Western blotting was used to measure the expression of DRP-1, OPA-1, MFN-1, HSP60, and SDHA in kidney tissues. LPS-induced sepsis mice were transfected with oe-negative control (NC) or oe-ITCH (ITCH overexpression). **p < 0.01.
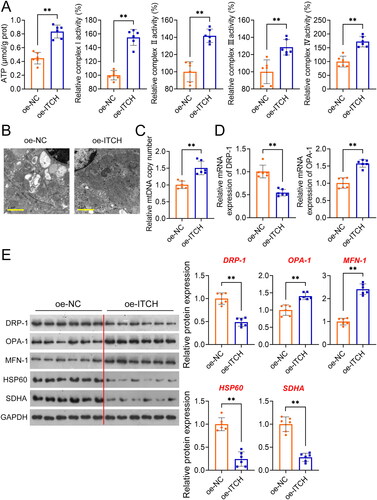
Figure 3. Circ-ITCH prevents apoptosis and inflammation in lipopolysaccharide (LPS)-stimulated HK-2 cells. (A) RT-qPCR was used to determine the relative expression of circ-ITCH in cells. (B) CCK-8 assay was used to detect cell viability. (C) Flow cytometry was used to identify cell apoptosis. (D) H2DCFDA probe was used to detect the reactive oxygen species (ROS) level in cells (scale bar = 25 µm). (E) Levels of malondialdehyde (MDA), superoxide dismutase (SOD), interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α in cells were detected by corresponding ELISA kits. *p < 0.05, **p < 0.01 vs. control; #p < 0.05, ##p < 0.01 vs. LPS + vector.
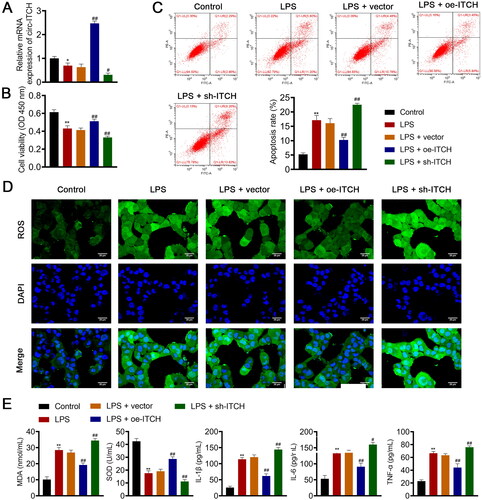
Figure 4. Circ-ITCH alleviates mitochondrial dysfunction in lipopolysaccharide (LPS)-stimulated HK-2 cells. (A) OCR was detected in cells. (B) RT-qPCR was used to determine the relative expression of DRP-1 and OPA-1 in cells. (C) Western blotting was used to measure the expression of DRP-1, OPA-1, and MFN-1 in cells. LPS- stimulated HK-2 cells were transfected with oe-ITCH (ITCH overexpression) or sh-ITCH (ITCH knockdown). **p < 0.01 vs. control; #p < 0.05, ##p < 0.01 vs. LPS + vector.
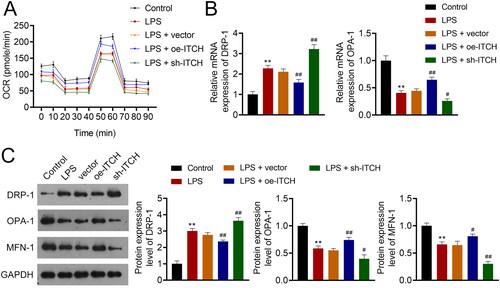
Figure 5. Circ-ITCH ameliorates mitochondrial dysfunction induced by oxidative stress in lipopolysaccharide (LPS)-stimulated HK-2 cells. (A) Levels of reactive oxygen species (ROS), malondialdehyde (MDA), and superoxide dismutase (SOD) in cells were detected by corresponding ELISA kits. (B) MitoSOX Red reagent was used to measure mitochondrial ROS in cells (scale bar = 25 µm). (C) Mitochondrial membrane potential of cells was monitored using JC-1 staining (scale bar = 25 µm). LPS-stimulated HK-2 cells were pretreated with N-cetylcysteine (NAC; a ROS scavenger) and transfected with sh-ITCH or sh-NC. **p < 0.01 vs. control; #p < 0.05, ##p < 0.01 vs. NAC + sh-NC.
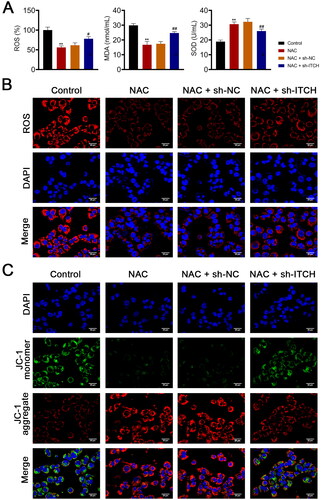
Figure 6. Circ-ITCH protects mitochondrial dysfunction induced by oxidative stress in lipopolysaccharide (LPS)-stimulated HK-2 cells. (A) Mitochondrial membrane potential (MMP) of cells was monitored using flow cytometry. (B) RT-qPCR was used to determine the relative expression of DRP-1 and OPA-1 in cells. (C) Western blotting was used to measure the expression of DRP-1, OPA-1, and MFN-1 in cells. LPS-stimulated HK-2 cells were pretreated with N-cetylcysteine (NAC; a ROS scavenger) and transfected with sh-ITCH or sh-NC. **p < 0.01 vs. control; #p < 0.05, ##p < 0.01 vs. NAC + sh-NC.
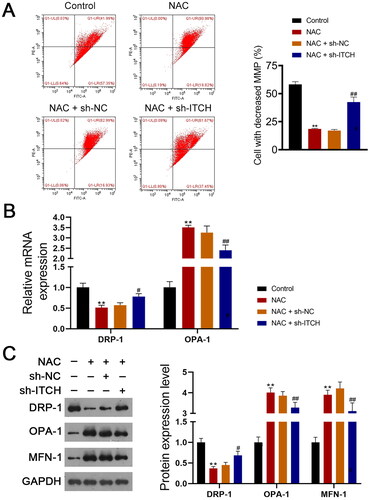
Figure 7. Circ-ITCH targets miR-214-3p/ABCA1 axis. (A) Circ-ITCH was identified to Localize to both nuclei and cytoplasms in HK-2 cells by fluorescence in situ hybridization (FISH) assay (scale bar = 25 µm). (B, C) the interaction between circ-ITCH and miR-214-3p was predicted using the starBase v2.0 database and identified by dual-luciferase reporter assay. (D, E) The interaction between miR-214-3p and ABCA1 was predicted using the starBase v2.0 database and identified by dual-luciferase reporter assay. (F, G) RT-qPCR was used to determine the relative expression of miR-214-3p and ABCA1 in kidney tissues of lipopolysaccharide (LPS)-induced sepsis mice with/without oe-ITCH transfection. (H) Western blotting was used to measure the expression of ABCA1 in LPS-induced sepsis mice. **p < 0.01.
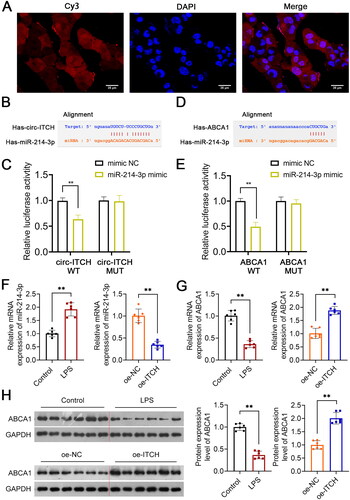
Figure 8. Circ-ITCH alleviates mitochondrial dysfunction of lipopolysaccharide (LPS)-stimulated HK-2 cells by targeting miR-214-3p/ABCA1 axis. (A) CCK-8 assay was used to detect cell viability. (B) Flow cytometry was used to identify cell apoptosis. (C) H2DCFDA probe was used to detect the reactive oxygen species (ROS) level in cells (scale bar = 25 µm). (D) Levels of malondialdehyde (MDA), superoxide dismutase (SOD), interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α in cells were detected by corresponding ELISA kits. (E) RT-qPCR was used to determine the relative expression of DRP-1 and OPA-1 in cells. (F) Western blotting was used to measure the expression of DRP-1, OPA-1, and MFN-1 in cells. (G) LPS-stimulated HK-2 cells were transfected with miR-214-3p inhibitor or/and sh-ITCH. **p < 0.01 vs. NC inhibitor; #p < 0.05, ##p < 0.01 vs. miR-214-3p inhibitor + sh-NC.
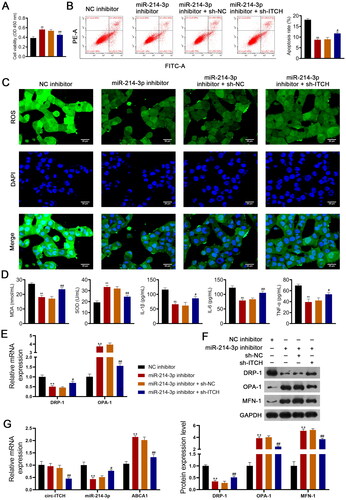
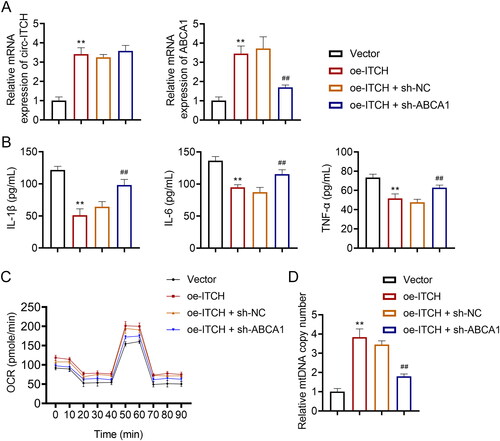
Figure 9. Regulatory interplay between circ-ITCH and ABCA1 in LPS-stimulated HK-2 cells. (A) RT-qPCR was used to determine the relative expression of circ-ITCH and ABCA1 in cells. (B) Levels of interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α in cells were detected by corresponding ELISA kits. (C) OCR was detected in cells. (D) RT-qPCR was used to determine the mtDNA copy number in cells. **p < 0.01 vs. vector; ##p < 0.01 vs. oe-ITCH + sh-NC.
Supplemental Material_Figure 1
Download TIFF Image (13.2 MB)Supplemental Material_Figure 2
Download TIFF Image (8 MB)Supplemental Material_Figure 3
Download TIFF Image (5.6 MB)Supplemental Material_Figure 4
Download TIFF Image (1.8 MB)Supplemental Material
Download PDF (717.8 KB)Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
