Figures & data
Table 1. Detailed data information of the Mendelian randomization study.
Figure 1. Flowchart of the bidirectional two-sample Mendelian randomization study. The design hypotheses are that the genetic variants are associated with celiac disease traits, but not with confounders, and the genetic variants are associated with the risk of CKD and the eGFRcrea only through celiac disease traits. Similar hypotheses are applicable for the reverse MR analysis.
Assumption i: IVs are significantly associated with celiac disease at a genome-wide level; assumption ii: IVs must remain independent of any confounders; assumption iii: IVs solely influence CKD and eGFRcrea through celiac disease.
GV, genetic variants; CKD, chronic kidney disease; eGFRcrea, estimated glomerular filtration rate levels based on serum creatinine; IV, instrumental variables.
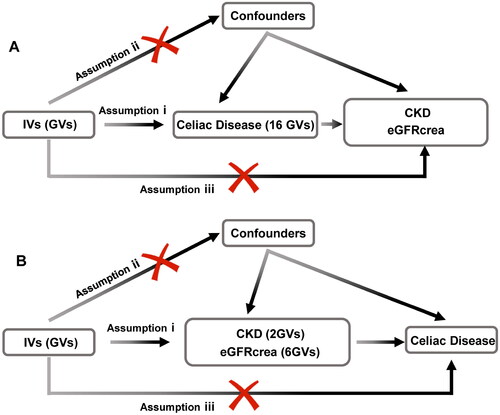
Table 2. Summary of genetic variants used to instrument celiac disease.
Figure 2. Scatter plots of Mendelian randomization. Scatter plot estimations of celiac disease as the expositional factor on CKD (A) and eGFRcrea (B). Each point in the scatter plot represents an IV. The line on each point reflects the 95% CI. A positive slope indicates that celiac disease had a positive effect on CKD. A negative slope indicates that celiac disease has a detrimental impact on eGFRcrea levels.
GV, genetic variants; CKD, chronic kidney disease; eGFRcrea, estimated glomerular filtration rate levels based on serum creatinine; IV, instrumental variables; CI, confidence interval.
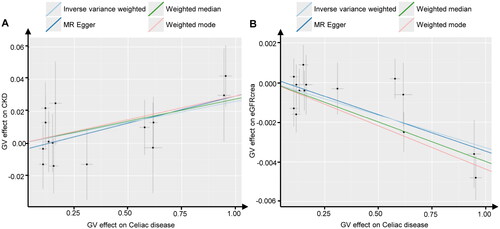
Table 3. Causal effects of bidirectional Mendelian randomization.
Figure 3. Leave-one-out tests of the genetic risks of celiac disease on CKD and eGFRcrea. The red lines are the analysis results of random effects IVW.
CKD, chronic kidney disease; eGFRcrea, estimated glomerular filtration rate levels based on serum creatinine; IVW, inverse variance weighted.
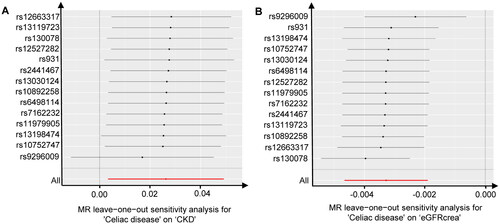
Table 4. Pleiotropy and heterogeneity tests of Mendelian randomization.
Supplemental Material
Download PDF (189.5 KB)Data availability statement
The data underlying this article were accessed from the IEU OpenGWAS consortium (https://gwas.mrcieu.ac.uk/).
