Figures & data
Figure 1. Identification of the potential DUBs of TLR4. (A) Venn diagram showing the potential DUBs predicted by Ubibrowser 2.0 software to bind TLR4, with overlapping proteins in three species: human (red), rat (purple), and mouse (green). (B) The expression levels of the above seven overlapping genes in renal tubule of sham and S-AKI groups were detected by western blot. Three biological replicates were performed in each group. (C–I) The protein expression correlation of TLR4 and seven overlapping genes was analyzed by Pearson correlation analysis. (J) Co-IP assay was used to verify the binding of Usp49, Stambp, Usp8, Usp9x, Usp33, Usp20, and Usp7 to TLR4. (K–L) Usp9x protein expression levels in LPS-treated NRK-52E cells were determined by Western blot, n = 3. NC was used as negative control, and β-actin was used as the internal reference protein. T-test was used to compare the difference between the two groups, **p < 0.01, compared with the NC group.
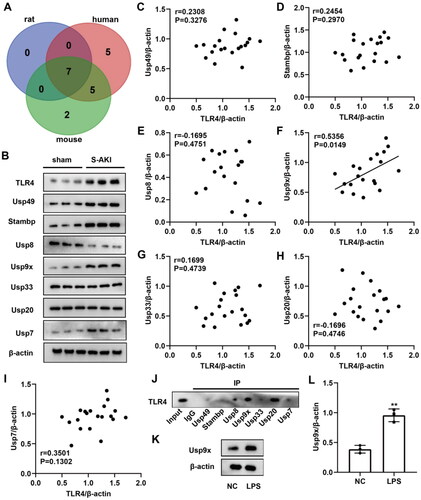
Figure 2. Usp9x enhances the stability of TLR4 protein through deubiquitination modification and promotes the TLR4/NF-κB signaling pathway. (A, B) The physical interaction between Usp9x and TLR4 was detected by Co-IP assay. (A) Cells were transfected with Flag-TLR4 plasmid for 48h. The cell lysate was co-immunoprecipitated with anti-Flag (IP: Flag), and then immunoblotting assay was performed with anti-Flag and anti-Usp9x. (B) Cells transfected with myc-Usp9x plasmid were co-immunoprecipitated with anti-myc (IP: myc) and then subjected to immunoblotting assay with anti-myc and anti-TLR4. (C) NRK-52E cells were co-transfected with Flag-TLR4, Usp9x-OV (or Vector), and HA-Ub. Cells were lysed and immunoprecipitation was performed with anti-Flag, and then immunoblotting assay was performed with anti-HA. (D-F) NRK-52E cell lines were transfected with control (Vector) or Usp9x overexpressed (Usp9x-OV) plasmid for 48h. (D) Cells were treated with 10 μg/mL CHX for intervals of 0-8h. The protein expression level of TLR4 was detected by western blot. β-actin is used as the internal reference protein. Two-way ANOVA (Bonferroni’s multiple comparisons test) was performed. *p < 0.05, **p < 0.01, compared to the Vector group. (E) NRK-52E cells were treated with or without 10 μM proteasome inhibitor MG132 for 4 h. TLR4 expression was detected by western blot. (F) The expressions of TLR4, p-IκB-α, Iκb-α, p-p65 and p65 in cells were detected by Western blot. β-actin is used as the internal reference of total protein. One-way ANOVA and Tukey’s multiple comparisons test were applied for multi-group comparison. The data are presented as the mean ± standard deviation (SD) of at least three independent experiments. **p < 0.01, ***p < 0.001.
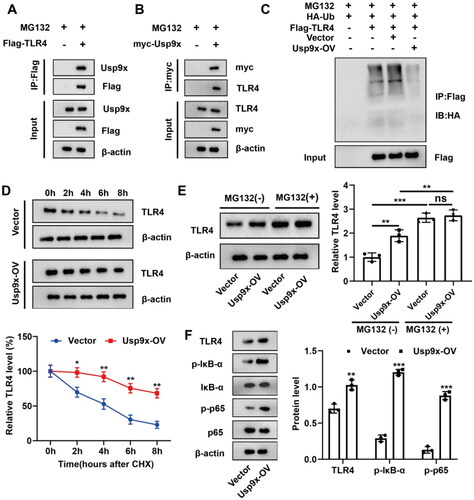
Figure 3. Interference with Usp9x inhibits inflammation and apoptosis of renal tubular epithelial cells. NRK-52E cells were transfected with LV-sh-Usp9x (sh-Usp9x) or LV-NC (sh-RNA) vectors for 48h, and then incubated with LPS (10 μg/mL) for 24h. (A) The expressions of Usp9x and TLR4 in cells were detected by western blot. The protein expression was analyzed statistically, n = 3. (B) The cells of each group were treated with MG132 (10 μM) for 4h before sample collection, followed by immunoprecipitation using anti-TLR4, and then anti-Ubiquitin was applied to detect the ubiquitination level of TLR4. (C-E) The levels of inflammatory cytokines IL-1β, IL-6, and TNF-α in the supernatant of NRK-52E cell lysate were detected by ELISA. (F) Apoptosis was detected by flow cytometry. (G) The results of apoptosis in (F) were statistically analyzed and three biological replicates were performed. (H) The expression of apoptotic proteins cleaved-caspase 3, caspase 3, Bax and Bcl-2 were detected by western blot, n = 3. (I) The expression of ER stress markers CHOP, ATF4, and GRP78 were detected by western blot, n = 3. One-way ANOVA (Tukey’s multiple comparisons test) was performed. **p < 0.01, ***p < 0.001, compared with the NC group; #p < 0.01, ##p < 0.01, ###p < 0.001, compared with LPS + sh-RNA group.
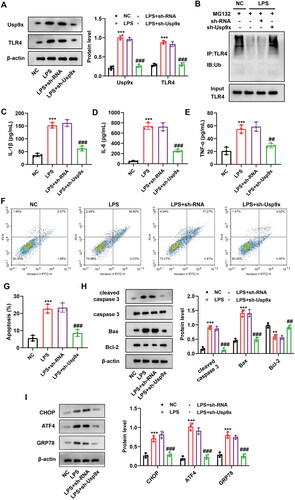
Figure 4. Interference with Usp9x alleviates septic acute kidney injury. Lentivirus carrying shRNA NC (LV-NC) or Usp9x shRNA (LV-sh-Usp9x) was injected into rats through the tail vein. CLP induction was performed two weeks after injection to construct S-AKI rat model. (A) The protein expressions of Usp9x, TLR4, p-IκB-α, IκB-α, p-p65, and p65 in renal tubules were detected by western blot, n = 3. β-actin is used as the internal reference protein. (B) The renal tubule lysates were IP with anti-TLR4, followed by IB with anti-Ubiquitin to analyze the ubiquitination modification of TLR4 protein. (C-I) Renal tubular biochemical indicators were measured by ELISA. One-way ANOVA (Tukey’s multiple comparisons test) was performed. (C-D) Scr and BUN levels; (E-F) Urine KIM1 and NGAL levels; (G-I) Serum levels of the inflammatory factors IL-1β, IL-6, and TNF-α. (J) HE staining of the renal tissues. The experiment was set up with three biological replicates. A representative image is shown in the figure. Bars = 50 μm. (K) TUNEL staining of the renal tissues. Bars = 50 μm. (L) Quantification of TUNEL-positive cells in each group. (M) The expression of ER stress markers CHOP, ATF4, and GRP78 were detected by western blot, n = 3. One-way ANOVA (Tukey’s multiple comparisons test) was used. ***p < 0.001, compared with sham group; ###p < 0.001, compared with S-AKI + LV-NC group.
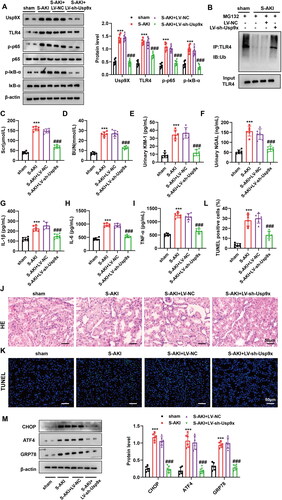
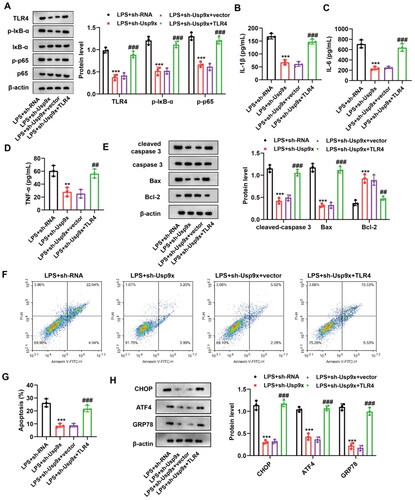
Figure 5. Interference with Usp9x alleviates renal tubular epithelial cell inflammation and apoptosis by inhibiting the TLR4/NF-κB pathway. (A) NRK-52E cells transfected with LV-sh-Usp9x (sh-Usp9x) or LV-NC (sh-RNA), or co-transfected with LV-sh-Usp9x (sh-Usp9x) and TLR4 overexpression vector (TLR4) or empty vector (vector) were treated with LPS. Cell lysates were then immunoblotted with the indicated antibodies (anti-TLR4, anti-p-IκB-α, anti-IκB-α, anti-p-p65, anti-p65, and anti-β-actin) to observe their expression. (B-D) Levels of the inflammatory factors IL-1β, IL-6, and TNF-α in each group were measured by ELISA. One-way ANOVA (Tukey’s multiple comparisons test) was used. (E) The expression of apoptotic proteins cleaved-caspase 3, caspase 3, Bax, and Bcl-2 were detected by western blot, n = 3. β-actin is used as the internal reference. (F) The apoptosis of each group was detected by flow cytometry. (G) The results of apoptosis rate in (F) were statistically analyzed. (H) The expression of ER stress markers CHOP, ATF4, and GRP78 were detected by western blot, n = 3. One-way ANOVA performed and statistically significant differences were indicated by: **p < 0.01, ***p < 0.001, compared with LPS + sh-RNA group; ##p < 0.01, ###p < 0.001, compared with LPS + sh-Usp9x + vector group.