Figures & data
Figure 1. Qualitative and quantitative analysis of biofilm formation conducted in a 24-well microtiter plate (modified from Lutskiy et al. Citation2015; Jung et al. Citation2018).
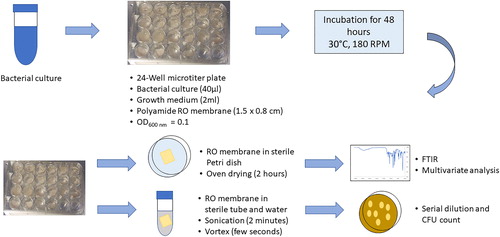
Figure 2. FTIR spectra of RO membranes after exposure to different media in the presence of H. aquamarina (incubation: 48 h at 30 °C; concentration: 1g l−1). Virgin RO – pure RO membrane surface; Negative control – MSM (no carbon source) with bacteria and MSM + carbon source (acrylic/maleic/poly acrylic acid/glucose) without bacteria; Positive control – glucose in MSM; AA + MSM – acrylic acid in MSM; PAA + MSM – poly acrylic acid in MSM; MA + MSM – maleic acid in MSM.
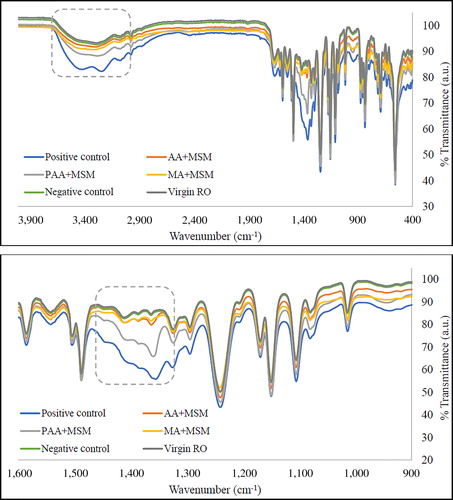
Table 1. Peak assignments to characterize biofilm layerTable Footnote*.
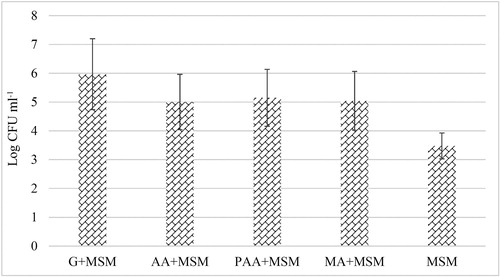
Figure 3. Increase in absorbance for selected peaks representing: (A) fatty acids and phospholipids, (B) polysaccharides, (C) protein components of the biofilm layer. Neg. control – MSM (no carbon source) with bacteria, and MSM + carbon source (acrylic/maleic/poly acrylic acid/glucose) without bacteria; Pos. control – glucose in MSM; AA + MSM – acrylic acid in MSM; PAA + MSM – poly acrylic acid in MSM; MA + MSM – maleic acid in MSM.
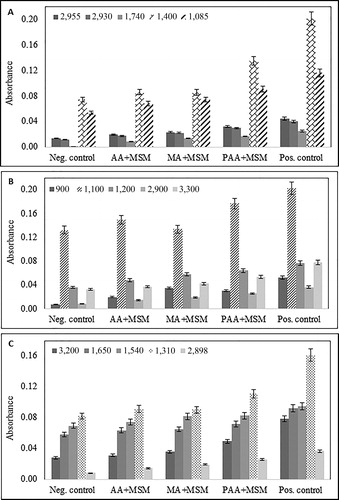
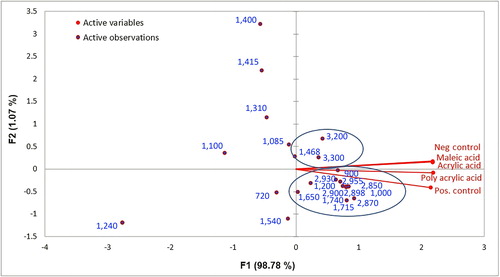
Figure 5. Biplot obtained for (A) the proteins; (B) the fatty acids and phospholipids; (C) the polysaccharide components of the biofilm using XLSTAT (V2016).
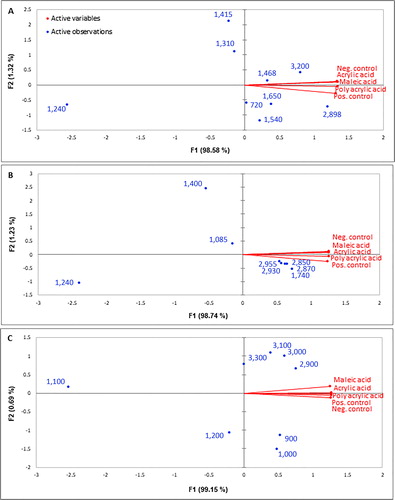
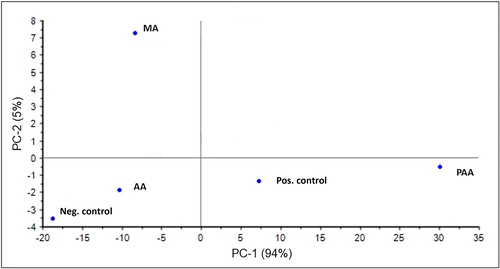
Figure 6. Clustering of the biofilm components obtained in the presence of different carbon sources.