Figures & data
Figure 2. The prediction error of the sum of the atomic energies for the 500 geometries in the test set.
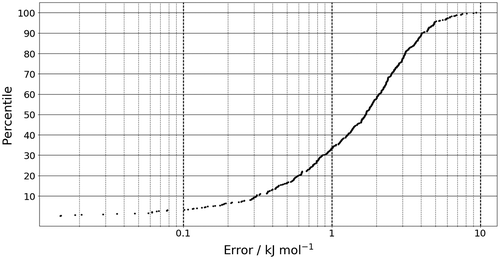
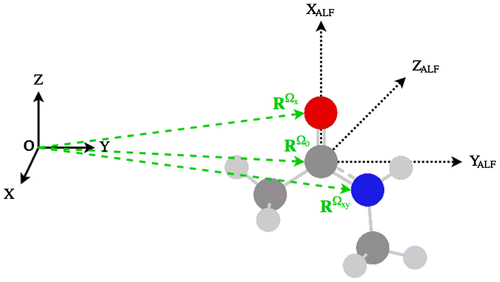
Figure 3. (Colour online) The global minimum geometry of peptide-capped glycine obtained at B3LYP/apc-1 level, with atomic labelling.
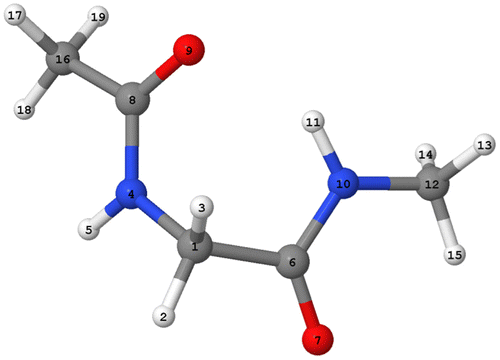
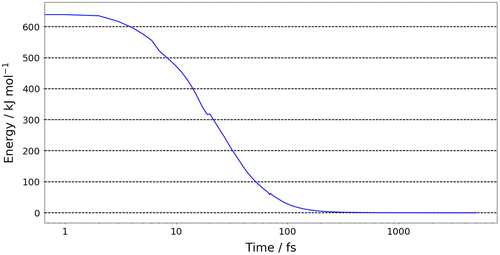
Figure 4. (Colour online) The energy convergence of 100 representative structures from the 4000 randomly generated structures, where the 0 kJ mol−1 values refers to the global minimum energy determined by GAUSSIAN09.
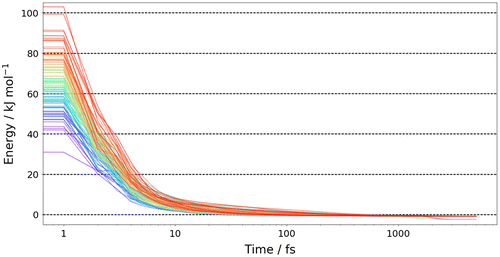
Table 1. A comparison of selected geometric properties of the global energy minimum generated by an ab initio calculation at the B3LYP/acp-1 level of theory and that generated by FFLUX.
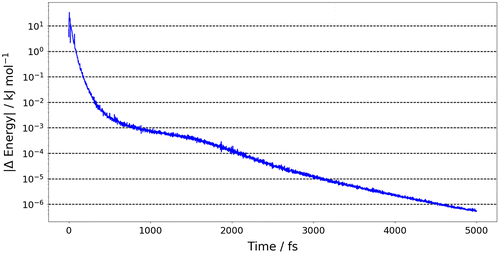
Figure 5. (Colour online) The relative energy (compared to the target minimum) of the system where the initial configuration was outside of the training domain as a function of time.