Figures & data

Fig. 1. CPY*-Inv escaped ERAD but underwent vacuolar degradation.
Note: (A) Schematic drawings of the fusion constructs used in this study. Invertase (Inv) is fused to the carboxyl termini of full-length CPY* and CPY to generate CPY*-Inv and CPY-Inv respectively. All constructs are expressed from the PRC1 promoter on a low-copy plasmid. (B) and (C) CHX chase assays were performed with wild-type (YKS12), hrd1∆ (YKS54), and pep4∆ (YKS28) cells expressing CPY* (B) or CPY*-Inv (C), as described in “Materials and methods.” The remaining proteins were analyzed by immunoblotting with anti-CPY antiserum and anti-3-phosphoglycerate kinase (PGK) antibody. Error bars represent standard deviations for three independent experiments. (D) Protein samples were prepared from the wild-type (YKS12, lanes 1 and 3) or pep4∆ cells (YKS28, lanes 2 and 4) harboring CPY-Inv (lanes 1 and 2) and CPY*-Inv (lanes 3 and 4) plasmid, treated with Endo H, and analyzed by immunoblotting with anti-CPY antiserum. (E) Protein samples were prepared from the pep4∆ cells (YKS28) expressing CPY* (lanes 1 and 2) or CPY*-Inv (lanes 3 and 4) with and without Endo H treatment, and were analyzed by immunoblotting with anti-CPY antiserum. (F) A CHX chase experiment for CPY*-Inv degradation in vps10∆ cells (YKS29) was done as described above.
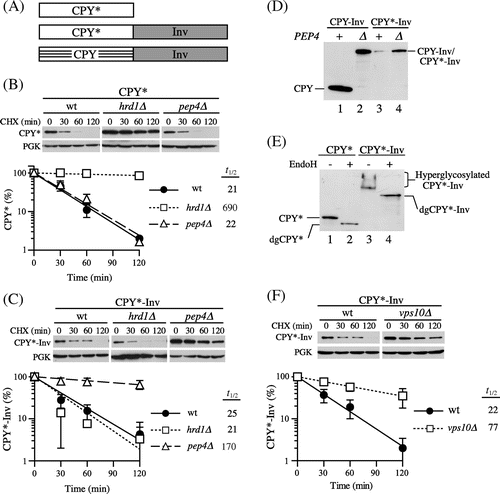
Fig. 2. CPY*-Inv but not CPY-Inv was sensitive to exogenously added protease.
Note: Cell lysates were prepared from prc1∆hrd1∆ pep4∆ (YKS30) cells expressing CPY (A), CPY* (A), CPY-Inv (B), or CPY*-Inv (B), and then incubated with or without trypsin for the indicated times, as described in “Materials and methods.” After Endo H treatment, samples were analyzed by SDS-PAGE, followed by immunoblot analysis with anti-CPY antiserum. Dots show tryptic digests, and arrowhead indicates the size corresponding to the CPY moiety.
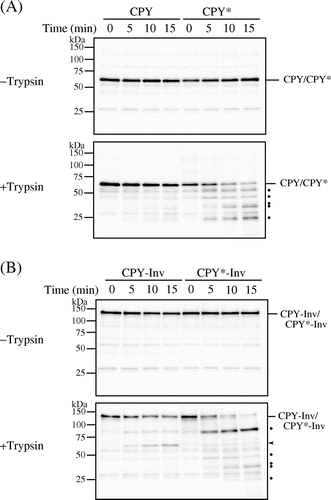
Fig. 3. Blockage of ER exit directed CPY*-Inv to ERAD.
Note: (A) Wild-type (YKS12), sec12-1 (YKS57), and hrd1∆sec12-1 (YKS61) cells containing CPY*-Inv plasmid were grown to early log phase at a permissive temperature. After incubation of the cells at a nonpermissive temperature for 30 min, CHX was added and aliquots were removed at the indicated times for immunoblot analysis, as described in Fig. (C). (B) Wild-type (YKS12), sec18-1 (YKS62), and hrd1∆sec18-1 (YKS78) cells containing CPY*-Inv plasmid were analyzed as described above.
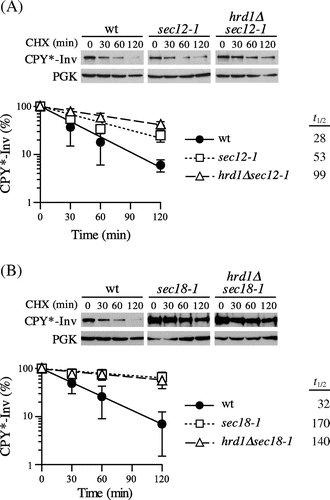
Fig. 4. Invertase domain in CPY*-Inv acted as an ER exit signal.
Note: (A) CHX chase experiments were run to monitor the degradation of CPY*-InvE486 K in the wild-type (YKS12), hrd1∆ (YKS54), and pep4∆(YKS28) cells, as described in Fig. (C). (B) pep4∆ cells (YKS28) expressing CPY*-Inv or CPY*-InvE486 K were grown to early log phase, and cell extracts were prepared. Samples were then analyzed by immunoblotting with anti-CPY antiserum.
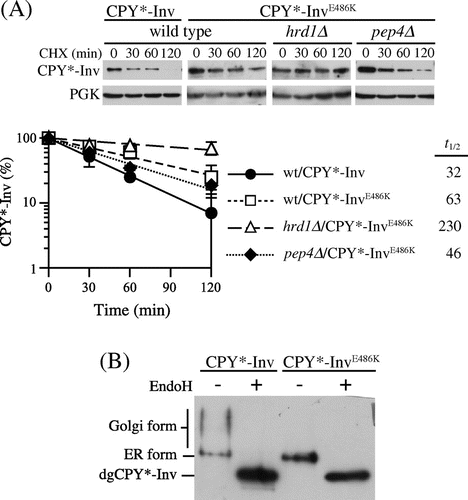
Fig. 5. CPY*-Inv escaped the ERAD via the p24 complex and Erv29.
Note: (A) CHX chase assays were performed in wild-type (YKS12), emp24∆ (YKS48), emp24∆hrd1∆ (YKS49), and emp24∆pep4∆ cells (YKS50) containing the CPY*-Inv plasmid. (B) CHX chase assays were performed in wild-type (YKS12), erv29∆ (YKS44), erv29∆hrd1∆ (YKS47), and erv29∆pep4∆ (YKS52) cells containing the CPY*-Inv plasmid. (C) CHX chase assays were performed in wild-type (YKS12), erv29∆emp24∆ (YKS59), hrd1∆erv29∆emp24∆ (YKS74), and pep4∆erv29∆emp24∆ cells (YKS75) containing the CPY*-Inv plasmid. (D) pep4∆ (YKS28), pep4∆erv29∆emp24∆ (YKS75), erv29∆pep4∆ (YKS52), and emp24∆pep4∆ cells (YKS50) expressing CPY*-Inv were grown to early log phase, and cell extracts were prepared. After the cell extracts were treated with Endo H or not treated, samples were analyzed by immunoblotting with anti-CPY antiserum.
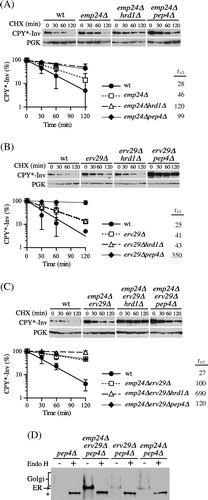
Fig. 6. The association of Kar2 and Yos9 with CPY*-Inv.
Note: (A) Microsomes were prepared from YTS346 cells expressing CPY-3HA, CPY*-3HA, CPY-Inv, or CPY*-Inv and subjected to native co-immunoprecipitation with anti-HA antibody. Microsomes (Input) and immunoprecipitates (IP: α-HA) were analyzed by immunoblotting with anti-CPY (IB: α-CPY) and anti-Kar2 (IB: α-Kar2) antisera and anti-myc antibody (IB: α-myc). (B) and (C) The relative amounts of Kar2 (B) and Yos9-myc (C) co-precipitated with CPY variants were quantified. The amount of Kar2 or Yos9-myc co-precipitated with CPY* was set to 1. Error bars represent standard deviations for three independent experiments.
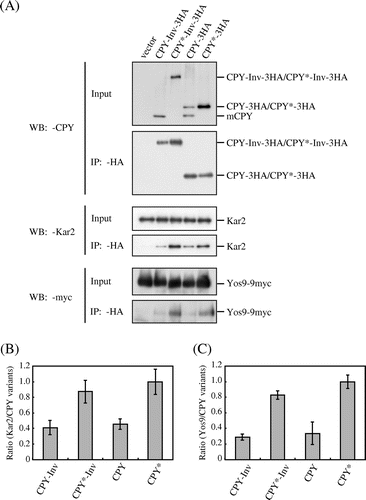
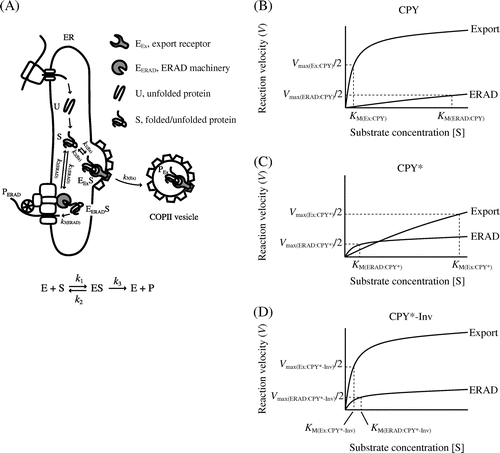
Fig. 7. Michaelis–Menten plots relate rates of ER export/ERAD to the concentrations of folded and misfolded secretory proteins.
Note: (A) A simplified FoldEx model for protein export. Unfolded (U) ensembles are converted to the folded or misfolded states (S) via ERAF. The S ensembles bind to ER export and ERAD machineries (EEx and EERAD, respectively) with specific rate constants (k1 and k2). Finally, the S ensembles are incorporated into COPII vesicles (PEx) or are retro-translocated to the cytosol (PERAD). The relationships between velocity of ER export or ERAD and the concentrations of folded or misfolded secretory proteins in the ER are adapted to Michaelis–Menten kinetics ((B) CPY; (C) CPY*; and (D) CPY*-Inv). Note that the graphs do not reflect actual values from experiments.