Figures & data

Table 1. Microbial strains used in this study.
Fig. 1. Succinic acid production in the SDH1- and SDH2-disrupted S. cerevisiae strains.
Note: (A) Cell growth, (B) glucose concentration, (C) succinic acid concentration, (D) ethanol concentration, (E) glycerol concentration, and (F) fumaric acid concentration in the BY4739 (hollow circles) and BY4739sdh12 (solid circles) strains are shown. Average ± standard deviation for three independent culture experiments is shown.
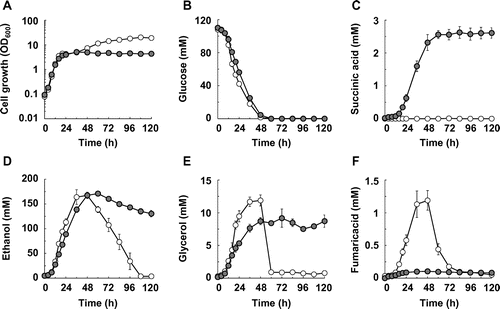
Table 2. Production yields in succinic acid-producing S. cerevisiae recombinant strains.
Fig. 2. Succinic acid production in ADH1–5-disrupted strain S149 and its SDH1- and SDH2-disrupted strain S149sdh12.
Note: (A) Cell growth, (B) glucose concentration, (C) succinic acid concentration, (D) ethanol concentration, (E) glycerol concentration, and (F) fumaric acid concentration in the S149 (hollow triangles) and S149sdh12 (solid triangles) strains are shown. Average ± standard deviation for three independent culture experiments is shown.
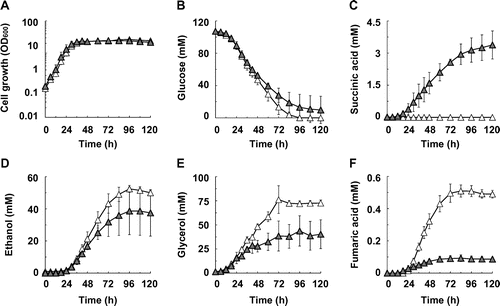
Fig. 3. Metabolic profiles of the S149 and S149sdh12 strains as analyzed by CE-TOFMS.
Note: The intracellular metabolite level (Y-axis, μmol g of dry cell−1) in the S149 (left, white bar) and S149sdh12 (right, gray bar) strains is shown. Average ± standard deviation for three independent culture experiments is shown. Asterisks indicate that the intracellular content was significantly different between the S149 and S149sdh12 strains. Glc, glucose; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde-3-phosphate; 1,3-BPG, 1,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; AcCoA, acetyl-CoA; AcAld, acetoaldehyde, EtOH, ethanol; PGL, phospho-glucono-1,5-lactone; 6PG, 6-phosphoglycerate; Ru5P, ribulose-5-phosphate; Xu5P, xylulose-5-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrose-4-phosphate; Cit, citrate; cis-Aco, cis-aconitate; IsoCit, isocitrate; 2OG, 2-oxoglutarate; SucCoA, succinyl-CoA; Suc, succinate; Fum, fumarate; Mal, malate; OAA, oxaloacetate.
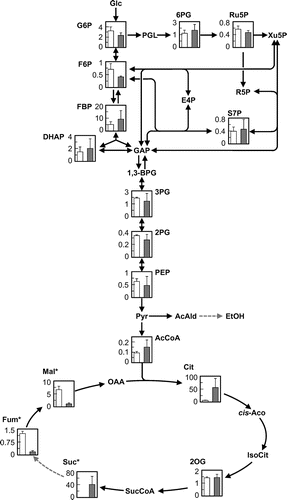
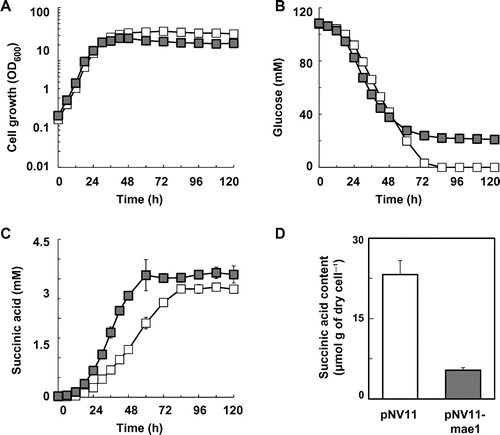
Fig. 4. Improvement of succinic acid production in the S149sdh12 strain by expression of the S. pombe mae1 gene.
Note: (A) Cell growth, (B) glucose concentration, (C) succinic acid concentration in the S149sdh12/pNV11 (hollow squares) and S149sdh12/pNV11-mae1 (solid squares) strains are shown. Intracellular succinic acid levels in the S149sdh12/pNV11 (white bar) and S149sdh12/pNV11-mae1 (gray bar) strains are shown (D). Average ± standard deviation for three independent culture experiments is shown.