Figures & data
Fig. 1. Transformation of C. merolae Cells.
Note: (A) Structure of the pKF-CG2 plasmid, which carries the URACm-Gs and APCC-GFP genes under their own promoters. It was constructed as follows: pCG2 plasmid DNA, which carries the APCC promoter (Papcc), the APCC (ORF ID: CMO250C)-GFP gene (Genbank ID: 332144728), and the polyadenylation signal sequence from the nopaline synthase gene (nos3′),Citation7,12) was digested with HindIII and treated with T4 DNA polymerase to make it blunt-ended. After digestion with EcoRI, the DNA fragment including Papcc, APCC-GFP, and nos3′ was purified. The plasmid vector DNA of pKFURACm-GsCitation13) was digested with SacI and treated with T4 DNA polymerase. After digestion with EcoRI, it was ligated with the APCC-GFP fragment. (B) Single-colony isolation from the transformant culture. The transformant culture (M4TF), made by introducing pKF-CG2 into the M4 strain, was diluted to the appropriate density and spotted on MA2 plates with embedded corn starch, and incubated for one month.
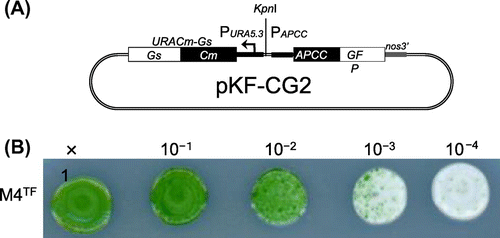
Fig. 2. Expression of the APCC-GFP Protein in C. merolae Cells.
Note: (A) Subcellular localization of APCC-GFP in the SG2 strain. Non-dividing (top) and dividing cells (bottom) were examined by fluorescence microscopy. From left to right, the images show phase-contrast microscopy, intrinsic chlorophyll fluorescence, fluorescence of GFP proteins, and superposition of chlorophyll and GFP fluorescence. Scale bar, 1 μm. (B) Immunoblot analysis with antiserum against GFP. Crude extracts were prepared from the C. merolae wild-type and SG2 cultures. Samples (20 μg for each lane), prepared as described previously,Citation6) were analyzed with mouse antiserum against GFP (MBL International, Boston, MA) as primary antibody and a horse radish peroxidase-conjugated anti-rabbit immunoglobulin G antibody (GE Healthcare Bio-Sciences) as secondary antibody.
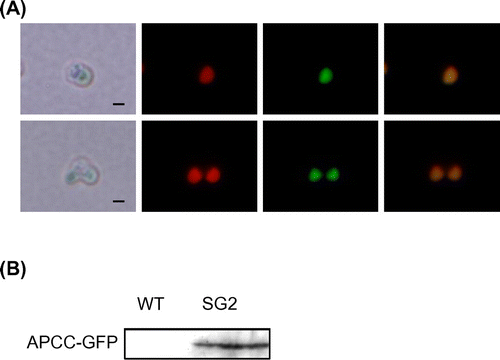
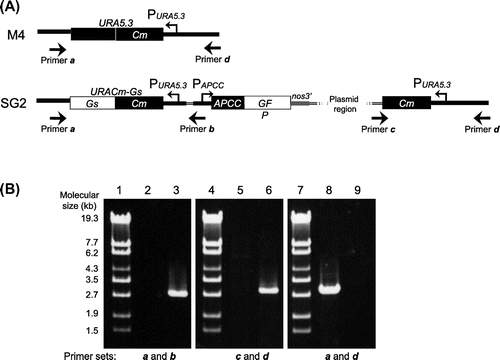
Fig. 3. Genomic Structure of the SG2 Strain.
Note: (A) Genomic structure around the URA5.3 gene in the M4 (top) and SG2 (bottom) strains. The annealing points of the primers are shown by arrows. (B) Agarose gel electrophoresis of the PCR products derived from the URA5.3 locus in the M4 (lanes 2, 5, and 8) and SG2 strains (lanes 3, 6, and 9). PCR was carried out with primer a (5-GCCGACGCTTGCTTCTGCCCATTAGG-3) and primer b (5-TCAAAAGACGGTGGTGCAGCGAGCGC-3) (lanes 2 and 3), primer c (5-ACGACGTTGTAAAACGACGGCCAGT-3) and primer d (5-ATGCACGGTGTCCAATAGGAGGAGGTCTCC-3) (lanes 5 and 6), and primers a and d (lanes 8 and 9) by ExTaq DNA polymerase (TaKaRa, Shiga, Japan). Lanes 1, 4, and 7, molecular size markers (λ-EcoT14 I digest).