Figures & data
Fig. 1. Lipid annotation scheme by means of the LipidBlast program.
Note: (A) Preparation of a hypothetical fragmentation pattern library. The LipidBlast program package (http://fiehnlab.ucdavis.edu/projects/LipidBlast/) contains the NIST Mass Spectral Search Program and a hypothetical MS/MS cleavage pattern library. For the hypothetical library, MS/MS cleavage rules of polar lipids were predicted based on experimentally obtained MS/MS fragmentation data on polar lipids (in-house data) and MS/MS fragmentation data on polar lipids from the published literature. The cleavage rules were applied to neutral and polar lipids (119,200 compounds). The hypothetical library consists of 78,314 positive MS/MS spectra (lipidblast-pos library) and 134,202 negative MS/MS spectra (lipidblast-neg library) with various adduct ions. (B) Execution of the LipidBlast program. Experimental MS/MS fragmentation data are used in a similarity search of the hypothetical MS/MS library. Neutral and polar lipid analyses are done by LC–MS/MS or infusion MS/MS. Analytical data (retention time, m/z and ion intensity information for parental ions, m/z and ion intensity information for MS/MS fragments) were converted into MGF files. For the LipidBlast program, the hypothetical MS/MS spectral library (lipidblast-pos or lipidblast-neg library) is installed into the NIST Mass Search Program. A similarity search by the LipidBlast yielded top hit lists of annotation information (lipid-head-group information, lipid carbon chain length information, lipid unsaturated bond number information, and detailed lipid structural information) with match scores for result reliability (Dot Product and Reverse Dot Product).
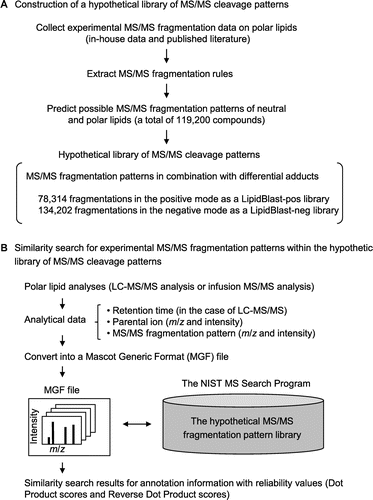
Fig. 2. Comparison of annotated lipid species of the E. gracilis Z strain before and after anaerobic culture.
Note: Nile Red staining images of E. gracilis Z strain with (a) and without (b) anoxia treatment (A). Cells were sampled at 0 h and 24 h after the initiation of anaerobiosis and stained with Nile Red solution.Citation18) Light microscopic images (left panel) and fluorescent microscopic images (right panel) are shown. Fluorescent images were visualized with an IX71 inverted microscope with a fluorescent imaging unit (Olympus, Tokyo). These Euglena cells were used in LC-IT/TOFMS analyses and LipidBlast profiling. Lipid classes of annotated lipid species (B), carbon chain lengths of annotated lipid species (C), and unsaturated bond numbers of annotated lipid species (D). Vertical bars compare the numbers of lipid annotations deduced from the experimental MS/MS fragmentation patterns on the basis of triplicate analyses. The lipid annotations were classified into lipid classes (shown below). In individual samples, the number of annotations belonging to each lipid class is shown in the percentage of the total annotation numbers (73, 76, and 73 annotations from the 0-h samples, and 94, 88, and 81 annotations from the 24-h samples). The error bars represent the standard deviations of triplicate experiments (independent sample extraction, MS analyses, and LipidBlast profiling). Asterisks indicate significant differences as compared to 0-h anoxia samples (*p < 0.05, **p < 0.01; Student’s t-test). White and gray bars indicate 0-h and 24-h anoxia treatments, respectively. The lipid classes are as follows: CerP, ceramide-1-phosphates; DG, diacylglycerols; lysoPC, lysoglycerophosphocholines; lysoPE, lysoglycerophosphoethanolamines; MG, monoacylglycerols; MGDG, monogalactosyldiacylglycerols; PA, glycerophosphates; PC, glycerophosphocholines; PE, glycerophosphoethanolamines; plasmenyl-PC, plasmenyl-glycerophosphocholines; plasmenyl-PE, plasmenyl-glycerophosphoethanolamines; PS, glycerophosphoserines; SM, sphingomyelins; and TG, triacylglycerols. NA not assigned.
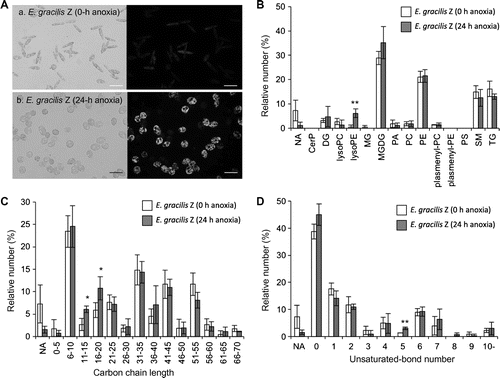
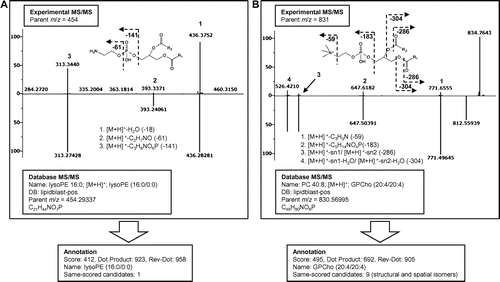
Fig. 3. Spectral similarity search results by the LipidBlast program (the library for positive mode analyses).
Note: Experimental MS/MS fragmentation data and retrieved hypothetical MS/MS fragmentation data are shown in the upper and lower panels, respectively. (A) A similarity search yielded the annotation lysoglycerophosphoethanolamine (lysoPE, 16:0/0:0) for experimental parent ion m/z 454. The annotation is shown with Dot Product and Reverse Dot Product values of 923 and 958, respectively. (B) A similarity search for the experimental parent ion m/z 831 yielded glycerophosphocholine (PC, 20:4/20:4) with the Dot Product and Reverse Dot Product values of 692 and 905, respectively. For this annotation, nine candidates with the same scores were generated due to the different positions of the double bonds.