Figures & data
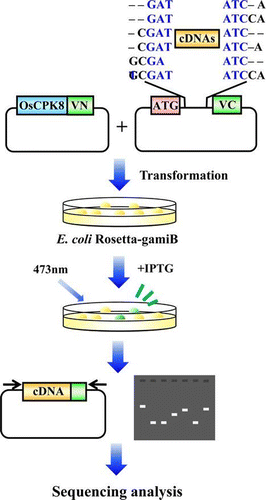
Fig. 1. BiFC analysis of interaction between OsMEK1 and OsMAP1 or OsMAP2 in rice protoplasts.
Note: (I) BiFC; (II) DsRed; (III) Merged image of BiFC, DsRed, and bright field. OsMEK1 was fused with the N-terminal fragment of Venus (OsMEK1-Vn). OsMAP1 and OsMAP2 were fused with the C-terminal fragment of Venus (OsMAP1-Vc and OsMAP2-Vc). OsMAP1-Vc, OsMAP2-Vc, and OsMEK1-Vn: each construct was individually co-transformed into rice protoplasts with DsRed expression vector. OsMEK1-Vn/OsMAP1-Vc: OsMEK1-Vn and OsMAP1-Vc were co-transformed into protoplasts with DsRed expression vector. OsMEK1-Vn/OsMAP2-Vc: OsMEK1-Vn and OsMAP2-Vc were co-transformed into protoplasts with DsRed expression vector. The transformed protoplasts were observed 12 h after the introduction of the vectors. Bar, 10 μm.
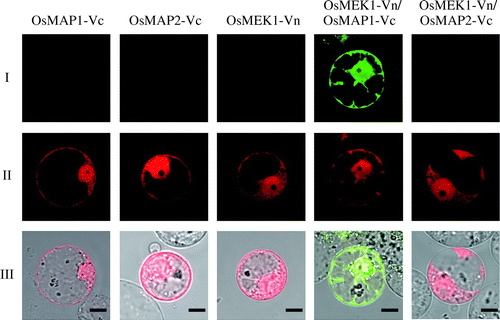
Fig. 2. Vectors used in the establishment of the screening system utilizing E. coli Rosetta-gami B.
Note: (A) Venus expression vector. pETTp-Venus contains the Venus gene placed between the T7 promoter and the T7 terminator. (B) OsMEK1-Vn expression vector. pETTp-OsMEK1-Vn contains OsMEK1 fused to the N-terminal fragment of the Venus gene (Vn) placed between the T7 promoter and the T7 terminator. (C) OsMAPs-Vc and GUS-Vc expression vectors. pCDF-X-Vc contains OsMAP1, OsMAP2, or OsMAP3 fused to the C-terminal fragment of Venus (Vc) placed between the T7 promoter and the T7 terminator. pCDF-GUS-Vc encodes GUS fused to the C-terminal fragment of Venus (Vc) placed between the T7 promoter and the T7 terminator. TmpR, trimethoprim resistance gene; SmR, streptomycin/spectinomycin resistance gene; GUS, β-glucuronidase.
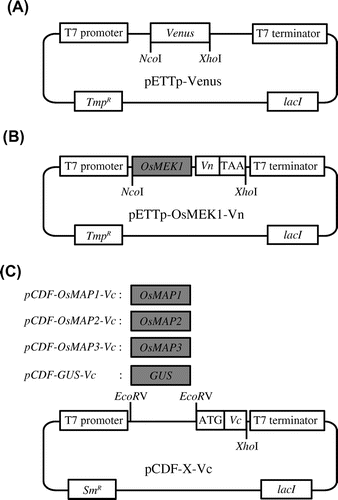
Fig. 3. Detection of specific interaction by BiFC in E. coli strain Rosetta-gami B.
Note: (A) Detection of fluorescence derived from BiFC of a culture plate. The cells contained the following vectors: 1, pETTp-Venus; 2, pETTp-Vn and pCDF-OsMAP1-Vc; 3, pETTp-OsMEK1-Vn and pCDF-OsMAP1-Vc; 4, pETTp-OsMEK1-Vn and pCDF-OsMAP2-Vc; 5, pETTp-OsMEK1-Vn and pCDF-OsMAP3-Vc; and 6, pETTp-OsMEK1-Vn and pCDF-GUS-Vc. Transformed E. coli Rosetta-gami B were cultured on nylon membranes left LB plates for 16 h at 37 °C, and then cultured on LB plates with 12.5 mm IPTG for 6 h at 25 °C. BiFC fluorescence was detected by means of an FLA3000 at an emission wavelength of 530 nm. (B) Immunoblot analysis of accumulated proteins in transformed cells by means of the anti-GFP antibody. The cells contained the following vectors: 1, pETTp-Venus; 2, pETTp-Vn and pCDF-OsMAP1-Vc; 3, pETTp-OsMEK1-Vn and pCDF-OsMAP1-Vc; 4, pETTp-OsMEK1-Vn and pCDF-OsMAP2-Vc; 5, pETTp-OsMEK1-Vn and pCDF-OsMAP3-Vc; and 6, pETTp-OsMEK1-Vn and pCDF-GUS-Vc. The same amount of each of the fractions was separated by SDS-PAGE, and proteins were detected by silver staining (below). Non-specific bands are indicated by asterisks.
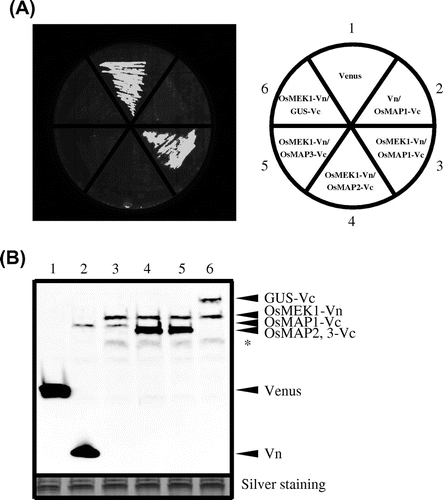
Fig. 4. Immune responses induced by OsCPK8 in rice.
Note: (A) Time-course of OsCPK8 expression in cultured rice cells following inoculation with the avirulent N1141 strain (hollow circle) and with the virulent K1 strain (solid circle) of A. avenae. The amounts of OsCPK8 mRNA were measured by real-time RT-PCR. Each data point represents the average of three independent experiments. Bars indicate standard deviation. (B) Enhancement of OsWRKY expression activity by transient overexpression of OsCPK8. Protoplasts isolated from cultured rice cells were transformed with OsWRKY70:: Luc, RLuc, and either empty vector or OsCPK8/pHAC17. OsWRKY promoter activities are represented by the ratio between firefly luciferase and Renilla luciferase activities, normalized against the value obtained for mock treatment. Asterisks indicate a significant increase (t-test; p < 0.05).
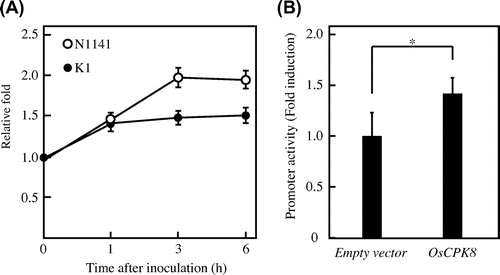
Fig. 5. BiFC screening for the detection of OsCPK8-interacting protein.
Note: (A) Vectors used in the rice cDNA-Vc library. pCDF-Vc-mix vectors contain six reading frames, and cDNAs were inserted into the EcoRV site. (B) OsCPK8-Vn expression vector. pETTp-OsCPK8-Vn contains OsCPK8 fused to the N-terminal fragment of Venus (Vn) placed between the T7 promoter and the T7 terminator. TmpR, trimethoprim resistance gene; SmR, streptomycin/spectinomycin resistance gene. C, Retest of specific interaction between OsCPK8 and CS1, CS2, and CS3. pETTp-OsCPK8-Vn was co-introduced into E. coli Rosetta-gami B along with pCDF-CS1-Vc, pCDF-CS2-Vc, pCDF-CS3-Vc, or pCDF-Vc, and the transformants were streaked on nylon membranes placed on LB plates with IPTG. After 6 h, bright fluorescence, derived from BiFC, was analyzed by illumination under a stereo fluorescence microscope.
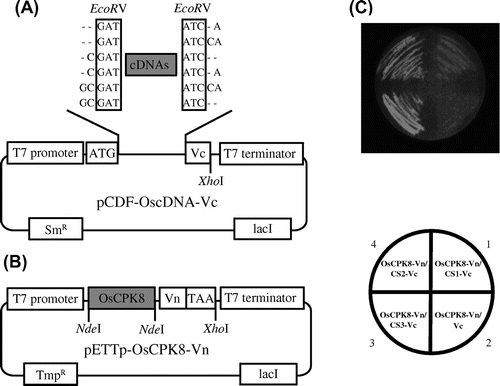
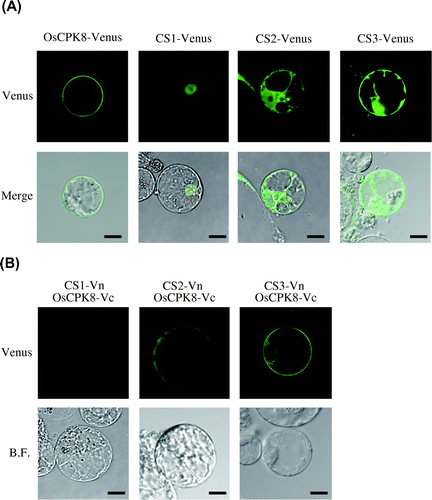
Fig. 6. BiFC analysis of the interaction between OsCPK8 and CS1, CS2, or CS3 in rice protoplasts.
Note: (A) Localization of the identified proteins in rice protoplasts. Venus-fused identified proteins (CS1-Venus, CS2-Venus, and CS3-Venus) were expressed in rice protoplasts. Upper panels are Venus images, and lower panels are merges of Venus and bright-field images. (B) Interaction between OsCPK8 and the identified proteins detected by BiFC in rice protoplasts. Proteins fused to the N-terminal fragment of Venus (Vn) (CS1-Vn, CS2-Vn, and CS3-Vn) and the C-terminal fragment of Venus (Vc) (OsCPK8-Vc) were co-expressed in rice protoplasts. Reconstructed Venus fluorescence and bright field were obtained by fluorescence confocal microscopy. Bar, 10 μm. BF, bright field.