Figures & data

Fig. 1. Structure of the lipid-linked oligosaccharide.
Notes: The Man residues added by Alg11p are underlined in bold. ManT-4 and ManT-5 are mannosyltransferase IV and mannosyltransferase V, respectively. The mannose residue added by each mannosyltransferase is underlined with a narrow line.
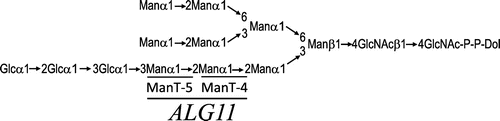
Fig. 2. Typical mouse EST clones homologous to the S. cerevisiae ALG11 gene.
Notes: EST clones BB64514 and BV703635 showed a 130-bp-gap and a 122-bp-gap, respectively.▼ indicates the site of GTAG insertion. Ten of 24 clones had the extra GTAG insertion.
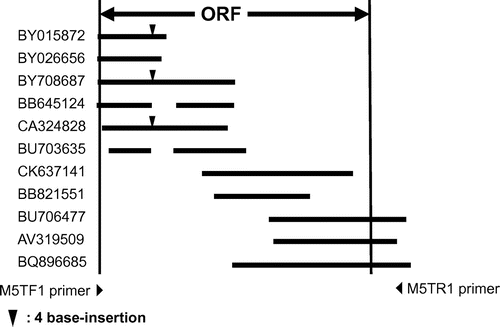
Fig. 3. Electropherogram of the ORF (270–303) of mALG11- and its variant-cDNA.
Note: Black arrow indicates the site of GTAG insertion in the variant cDNA.
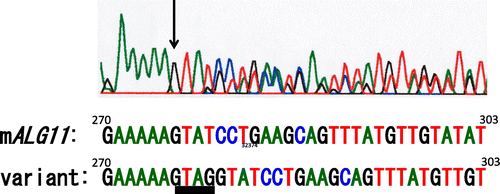
Fig. 4. ClustalW sequence alignment of the predicted amino acid sequences of mouse-, human-, C. elegans-, S. pombe-, and S. cerevisiae- mannosyltransferase IV/V.
Symbols: (*) identical residues; (:) conserved substitutions; (.) semi-conserved substitutions. Transmembrane regions are boxed.

Fig. 5. Possible mechanism of GTAG insertion in the variant of mALG11 cDNA.
Notes: Partial nucleotide sequences of the exon 2–intron 2 boundary region and the intron 2–exon 3 boundary region of mouse genomic clone AC117665 are shown. Black letters indicate nucleotides in the 3′-terminal region of exon 2 and the 5′-terminal region of exon 3, and gray letters indicate nucleotides in the 5′-terminal region and the 3′-terminal region of intron 2. The variant cDNA contains an extra four nucleotide insertion derived from 5′ end of intron 2 of mALG11 genomic DNA.

Fig. 6. Comparison of the genomic DNA structure and partial cDNA of sequences of mALG11 (A) and hALG11 (B).
Notes: Left panels show the exons (white boxes: exon 1 (1–44; 44 bp), exon 2 (45–275; 231 bp), exon 3 (276–490; 215 bp), and exon 4 (491–1476; 986 bp)), and introns (black boxes) those of mALG11 ORF and hALG11 ORF, which were obtained in our study and from the DDBJ/EMBL/GenBANK database. The nucleotide sequences (141–350) of the ORF of mALG11 and hALG11 cDNA are shown in the right panel. Underlined italics are non-identical nucleotides between the mALG11 and the hALG11 cDNA. Lines show the boundary region between exon 2 and exon 3 of mouse and of human ALG11 cDNA.
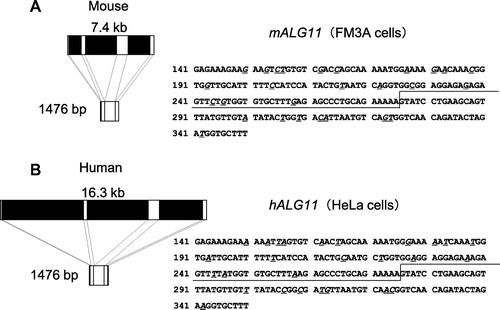
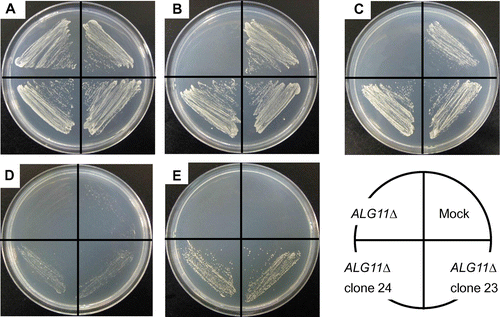
Fig. 7. Recovery of temperature-sensitive growth of the S. cerevisiae ALG11Δ Mutant by Transfection of mALG11 cDNA.
Notes: Parental ALG11Δ, mock-transfected ALG11Δ, and mALG11 (clones 23 and 24)-transfected ALG11Δ were plated on an SCD Trp+ agar plate and grown at 26 °C (A), plated on an SCD Trp- agar plate and grown at 26 °C (B), plated on an SCG Trp- agar plate and grown at 26 °C (C), plated on an SCD Trp- agar plate and grown at 37 °C (D), or plated on an SCG Trp- agar plate and grown at 37 °C (E).