Figures & data
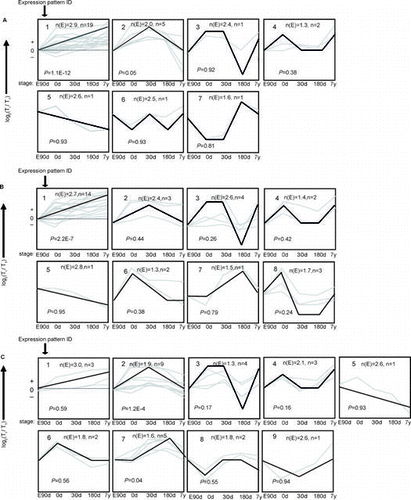
Table 1. The biological process enrichment of genes in significant expression pattern.
Fig. 1. The trends of 30 mRNA were divided into groups according to their dynamic expression patterns at the age of embryonic day 90 (E90d), postnatal day 0 (0d), day 30 (30d), day 180 (180d), and 7 years (7y) for (A) cardiac muscle, (B) longissimus dorsi Muscle, and (C) psoas major Muscle.
Notes: The dashed line indicates no change in expression at different age stages. The number in the top left corner of each square indicates the expression pattern ID. The blank, bold lines in the squares are trendlines of the expression patterns, and the gray lines represent gene expression from embryonic day 90 to 7 years postnatally. The p value is the corrected p value between the number of genes expected (n(E)) and the number of genes assigned (n), with p values < 0.05 considered to be statistically significant. Four other expressions were normalized to the highest one in four stages firstly, and then all expressions were log2-transformed.
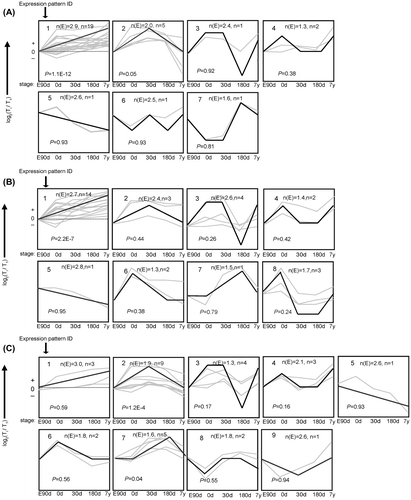
Fig. 2. Gene expression related to glycolysis in cardiac and skeletal muscle during development.
Notes: Triose phosphate isomerase 1 (TPI1), muscle phosphofructokinase (PFKM), and monocarboxylate transporter 4 (MCT-4) are important glycolytic enzymes that are mainly expressed in white muscle, and significantly up-regulated in LDM compared with CM and PMM in the study. The data were normalized to the highest expression of the three muscles and presented as mean ± SD. n = 3 for each age stages of tissues. CM, cardiac muscle; LDM, longissimus dorsi muscle; PMM, psoas major muscles.
**p < 0.01, *p < 0.05.
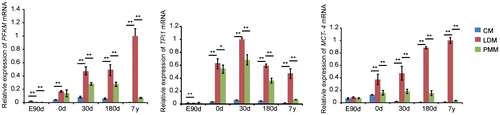
Fig. 3. Gene expression related to oxidation in cardiac and skeletal muscle during development.
Notes: ATPase-ß, citrate synthase (CS), and pyruvate dehydrogenase (PDH1) are related to mitochondrial oxidation, which were up-regulated in PMM compared with CM and LDM in this study. The data were normalized to the highest expression of the three muscles and presented as mean ± SD. n = 3 for each age stages of tissues. CM: cardiac muscle, LDM, longissimus dorsi muscle, PMM: psoas major muscle.
**p < 0.01, *p < 0.05.
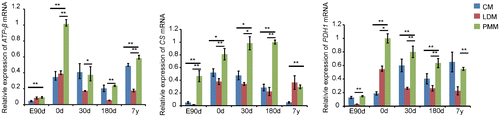
Fig. 4. The trends of 10 miRNA were divided into groups according to heir dynamic expression patterns for (A) Cardiac Muscle, (B) longissimus dorsi Muscle, and (C) psoas major muscles at the age of embryonic day 90 (E90d), postnatal day 0 (0d), day 30 (30d), day 180 (180d), and 7 years (7y).
Notes: The dashed line indicates no change in expression among different stages. The number in the top left corner of each square is the expression pattern ID. The blank, bold lines in the squares are trendlines of the expression patterns, and the gray lines represent gene expression from embryonic day 90 to 7 years postnatally. The p-value is the corrected P-value between the number of genes expected (n(E)) and the number of genes assigned (n), with p values < 0.05 considered to be statistically significant. Four other expressions were normalized to the highest one in four stages firstly, and then all expressions were log2-transformed.
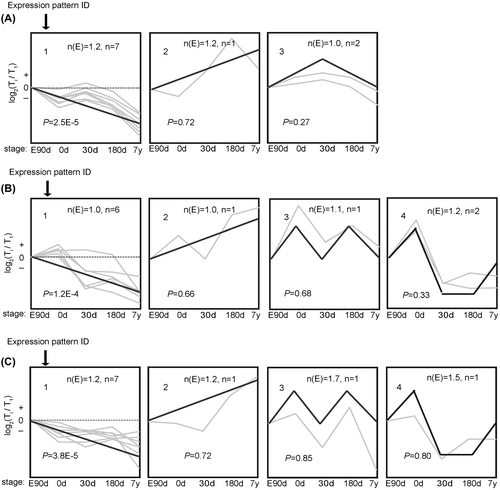
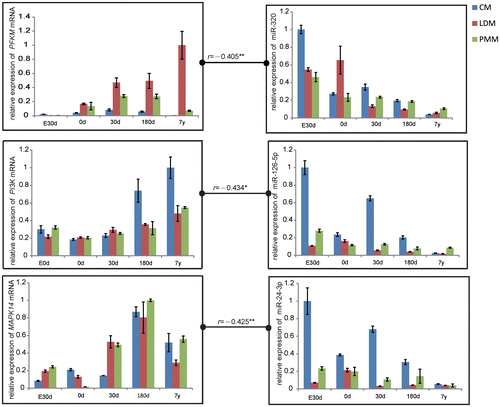
Fig. 5. Correlation in Gene Expression between PFKM and miR-320, PI3 K and miR-126, MAPK and miR-24 at the Stage of Embryonic day 90 (E90d), Postnatal day 0 (0 d), Day 30 (30d), Day 180 (180d), and 7 years (7y).
Notes: The data were normalized to the highest expression of the three muscles and presented as mean ± SD. n = 3 for each age stages of tissues. CM, cardiac muscle; LDM, longissimus dorsi muscle; PMM, psoas major muscles.
**p < 0.01, *p < 0.05.