Figures & data
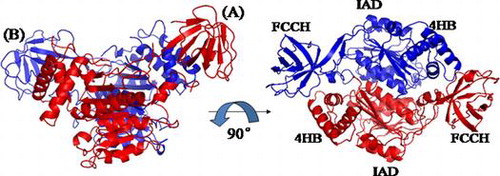
Fig. 1. Domain architectures of human and S. cerevisiae ubiquitin E1.
Notes: A, adenylation domain; CC, cysteine catalytic domain; UFD, ubiquitin-fold domain. C632 and C600 are the active site cysteine residues for human and S. cerevisiae E1, respectively.
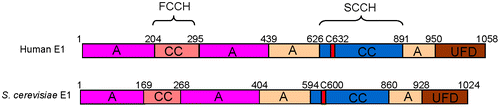
Fig. 2. Cloning of the N-terminal domains of human ubiquitin E1.
Notes: The N-terminal domains of human ubiquitin E1 were amplified and inserted into the pETDuet-1 vector with an N-terminal 6 × His tag for recombinant expression.
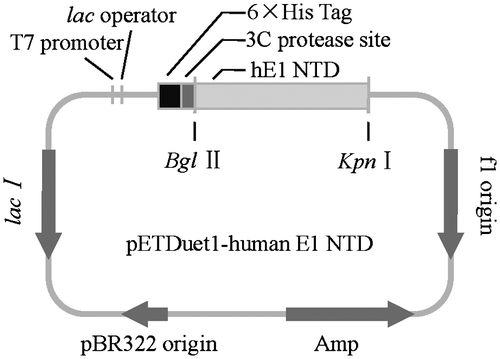
Fig. 3. SDS-PAGE of the purified N-terminal domains of human ubiquitin E1.
Notes: The purified proteins were resolved via 12% SDS-PAGE, stained with Coomassie Brilliant Blue R-250, and then de-stained with an ethanol: acetic acid: water (2:1:7) mixture. The left lane is the protein marker, and the right lane contains 1.5 μL of the purified recombinant proteins.
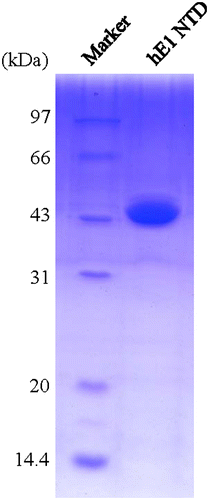
Fig. 4. Crystallization of N-terminal domains of human ubiquitin E1.
Notes: (A) Crystals of N-terminal domains of human ubiquitin E1 after three days at room temperature with the reservoir buffer containing 0.1 M Na3Citrate pH 5.6 and 3.2 M NH4Ac. (B) Optimized crystals of N-terminal domains of human ubiquitin E1 by microseeding, after two days growth at room temperature with the reservoir buffer containing 0.1 M Na3Citrate pH 5.6 and 3.0 M NH4Ac.
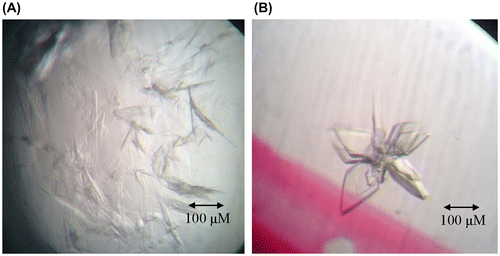
Fig. 5. Diffraction patterns of the crystal of the N-terminal domains of human ubiquitin E1.
Notes: The crystal diffracted to 2.75 Å resolution. B is the partial enlargement of region in A surrounded by rectangle.
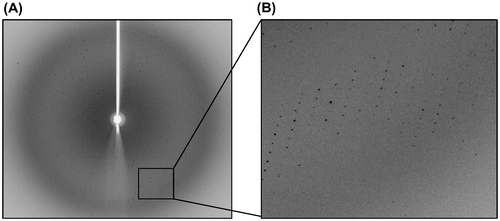
Table 1. Data collection and refinement statistics.
Fig. 6. Sequence alignment of the N-terminal domains of human ubiquitin E1 (1-449) and S. cerevisiae E1 (1-424).
Notes: Secondary structures of human ubiquitin E1 (1-439) and S. cerevisiae E1 (1-424) were labeled above and below the amino acid sequence based on the crystal structure (PDB ID: 4P22 and 3CMM).
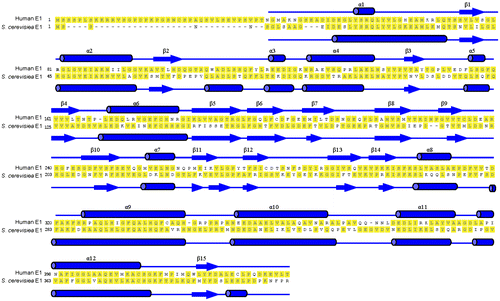
Fig. 7. Overall Structure of N-terminal domains of human ubiquitin E1.
Notes: Schematic diagram of the structure of the N-terminal domains of human E1, and the right figure is the view of the left structure by a 90° rotation around a horizontal axis. A and B are two molecules in the same crystallographic asymmetric unit.
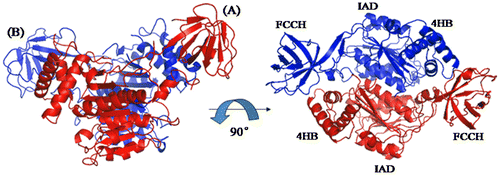
Fig. 8. Structural Comparison of the N-terminal domains of human ubiquitin E1 with Its Yeast Homologs.
Notes: (A and B) Superimposed N-terminal domains of human ubiquitinE1 (colored blue) with S. cerevisiae Uba1 1-424 (PDB ID: 3CMM, colored pink) and S. pombe Uba1 1-392 (PDB ID: 4II2, colored cyans). (C) Superimposed FCCH domains from human ubiquitin E1 and yeast Uba1 (the same color with A and B). (D, E, and F) The relative position of human, S. cerevisiae and S. pombe ubiquitin E1 FCCH domains with the IAD, the encircled region is the ubiquitin-binding surface.
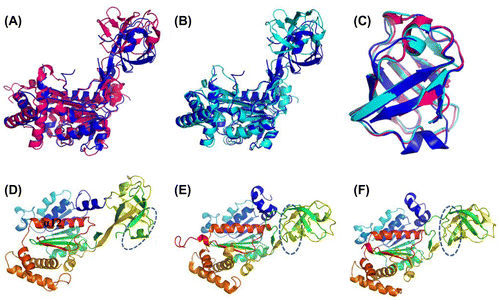
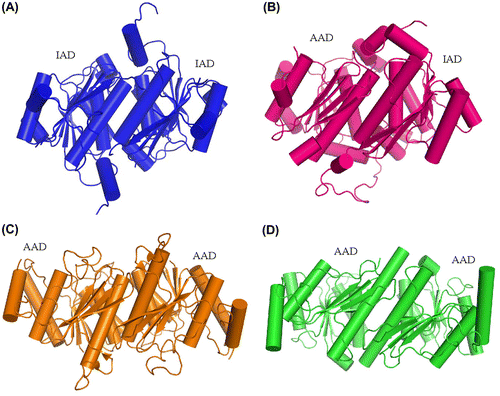
Fig. 9. Comparison of the dimerization pattern of human ubiquitin E1 IAD with Its homologs in yeast uba1 and non-canonical E1s.
Notes: (A) Cartoon diagram of human E1 IAD dimer in the same crystallographic asymmetric unit. (B) Pseudo-symmetrical dimerization of S. cerevisiae Uba1 IAD and AAD (PDB ID: 3CMM). (C and D) Dimerization of the Atg7 and Uba5 AADs, respectively (PDB ID: 3VH1 and 3H8 V).