Figures & data

Fig. 1. Comparison of amino acid sequences of hydrophobins EAS, RolA, and HFBII.
Notes: (A) Sequences of class I hydrophobins Aspergillus oryzae RolA and N. crassa EAS and class II hydrophobin T. reesei HFBII. The eight cysteine residues that form the conserved disulfide bonds are indicated in bold type. Selected hydrophobic amino acid residues of RolA for creating RolA mutants were highlighted by gray color. L137 and L142 of RolA were underlined. The alignment was generated with GENETYX (ver. 10). (B) Similarity scores of the amino acid sequences of RolA, EAS, and HFBII, calculated with GENETYX (ver. 10).
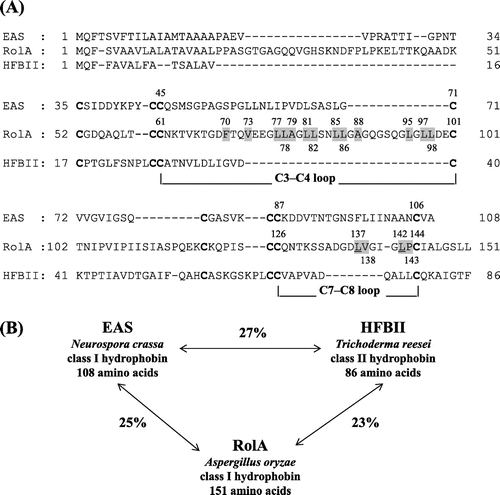
Fig. 2. Pull-down Assay of RolA mutants to PBSA microparticles.
Notes: Adsorption of RolA mutants in which a hydrophobic amino acid in the hydrophobic region was replaced with serine on PBSA microparticles was examined. PBSA microparticles were incubated with the culture supernatants of RolA mutant strains at 30 °C for 30 s. After the incubation, the RolA–PBSA complex was collected by centrifugation, and the proteins bound to the PBSA microparticles were extracted with SDS-PAGE loading buffer and subjected to SDS-PAGE. The data are means ± SD (n = 3).
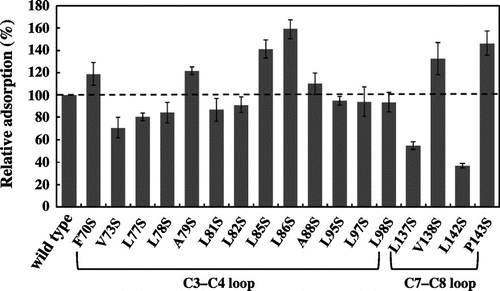
Fig. 3. CD spectra of RolA in the soluble form.
Notes: Wild-type RolA (solid line), RolA-L137S mutant (dotted line), RolA-L142S mutant (short-dashed line), and RolA-L137/L142S double mutant (long-dashed line). CD spectra are the averages of 10 scans collected by using a reference solution without the protein.
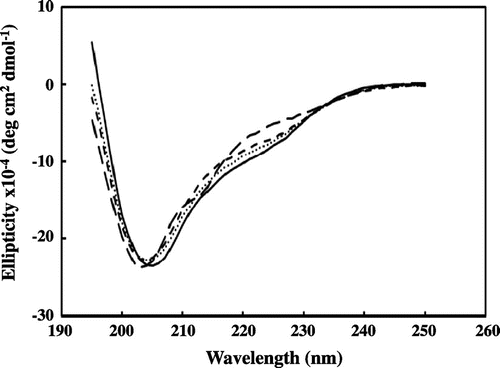
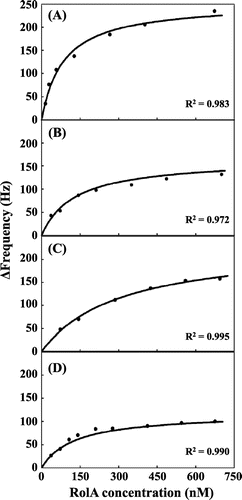
Fig. 4. QCM analysis of interaction of RolA and its mutants with PBSA.
Notes: To evaluate the affinity of binding of RolA and its mutants to PBSA, the frequency changes upon addition of RolA or its mutants to the QCM analysis chamber were analyzed by fitting to a Langmuir adsorption isotherm plot by using Aqua software (ver. 1.2, Initium). (A) wild-type RolA, (B) RolA-L137S mutant, (C) RolA-L142S mutant, and (D) RolA-L137S/L142S double mutant. R2 is the coefficient of determination.