Figures & data

Table 1. Primers used in this study.
Fig. 1. Production and purification of His6-MBF.
Notes: Purified recombinant His6-MBFs from LGG and FSMM22 were separated by SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Molecular mass standards are indicated on the left.
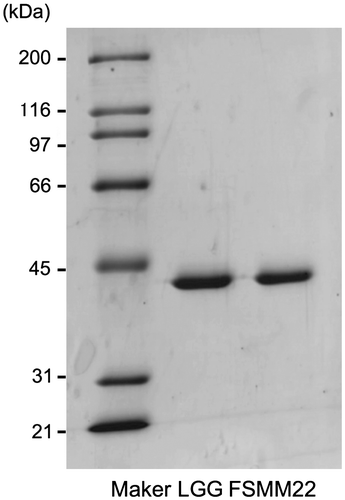
Fig. 2. Binding of His6-MBF from FSMM22 to PCM and ECM proteins.
Notes: (a) Overlay dot blot showing binding of His6-MBF to PCM, laminin (Ln), fibronectin (Fn), collagen IV (Cn IV), and BSA. Bound PCM and ECM proteins were detected using an anti-His tag antibody. (b) Biacore X sensorgrams of the interaction of His6-MBF with PCM, laminin, fibronectin, collagen IV, and BSA. His6-MBFs at the indicated concentrations were injected onto a CM5 sensor chip with immobilized PCM or ECM proteins. The measured data (black line) and their global fits are overlaid (red line). The KD value is indicated in the text and Table
Table 2. Affinity and rate constants for interactions of His6-MBF from FSMM22 with PCM and ECM proteins determined by SPR analysis.
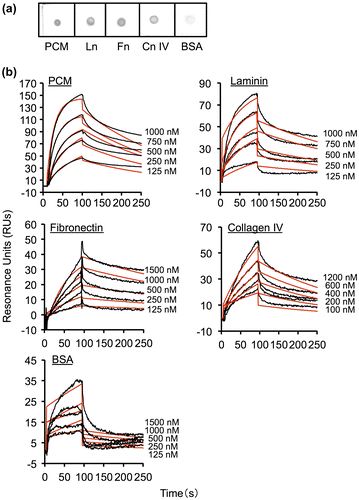
Table 2. Affinity and rate constants for interactions of His6-MBF from FSMM22 with PCM and ECM proteins determined by SPR analysis.
Fig. 3. Effect of different NaCl concentrations on His6-MBF binding.
Notes: Binding of His6-MBF (500 nM) from FSMM22 to immobilized PCM and ECM proteins was analyzed by SPR. HBS-EP buffer was supplemented with NaCl to a final concentration ranging from 150–450 mM. Results were expressed as relative percentage His6-MBF binding compared to 150 mM NaCl. Error bars indicate SD (n = 3).
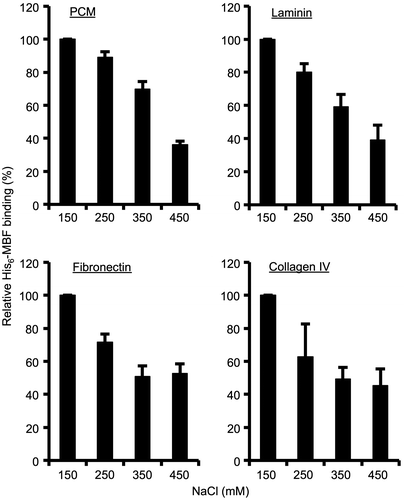
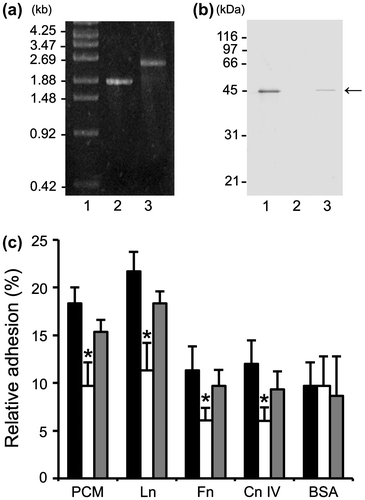
Fig. 4. Effect of mbf deletion on FSMM22 adhesion to PCM and ECM proteins.
Notes: (a) A genotype analysis of the resultant cmr gene into the genome-encoded mbf gene was performed by PCR using primer pair S1/S2. Amplified fragments were subjected to agarose gel electrophoresis. Lanes: 1, size maker; 2, wild-type strain; and 3, mbf mutant. The sizes of representative marker fragments are shown to the left (kb). (b) MBF was detected in the whole-cell lysates of FSMM22 wild-type strain (Lane 1), mbf mutant (Lane 2), and mbf-complemented strain (Lane 3) by Western blotting with an anti-MBF antibody. The position of the MBF protein is highlighted (arrow). The sizes of representative marker fragments are shown to the left (kDa). (c) Adhesion was examined for FSMM22 wild-type strain (black bar), mbf mutant (white bar), and mbf-complemented strain (gray bar) to PCM, laminin, fibronectin, collagen IV, and BSA. Asterisks indicate significant differences in binding (*p < 0.05) compared to wild-type strain, as analyzed by one-way ANOVA with the Dunnett’s post hoc test. (n = 5).